Tutorial
1. How to prepare the inputs for Alz-Disc prediction?
First, you need to prepare your interest of protein sequence in FASTA file format. For more information about FASTA fromat click here.
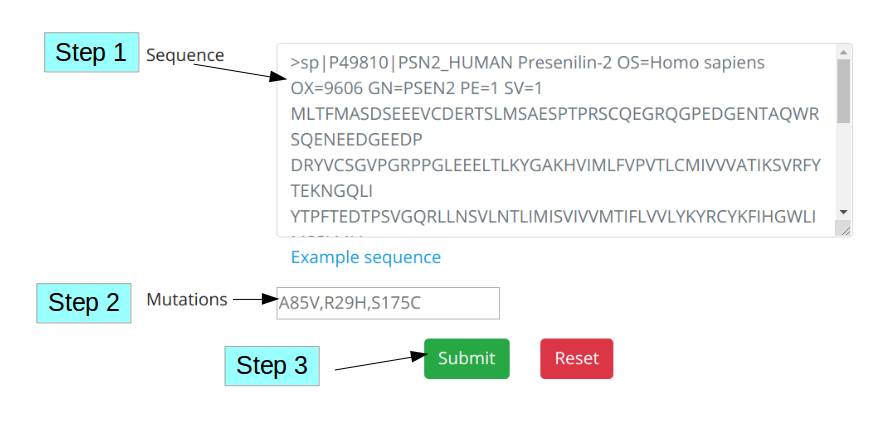
Then, paste your protein sequence in the sequence text box, which is given in the “Prediction” tab (Step 1 in the following figure).
Next, prepare your missense mutation using the following format:
Syntax: {Wild-type residue in single letter code}{position of wild-type residue according to the given protein sequence}{Name of the mutant residue in single letter code}
Example: A85V
After that, paste this information in the respective text box in the “Prediction” tab (Step 2 in the following figure). Finally, click submit button to start the Alz-Disc prediction (Step 3 in the following figure).
2. How to interpret the Alz-Disc output??
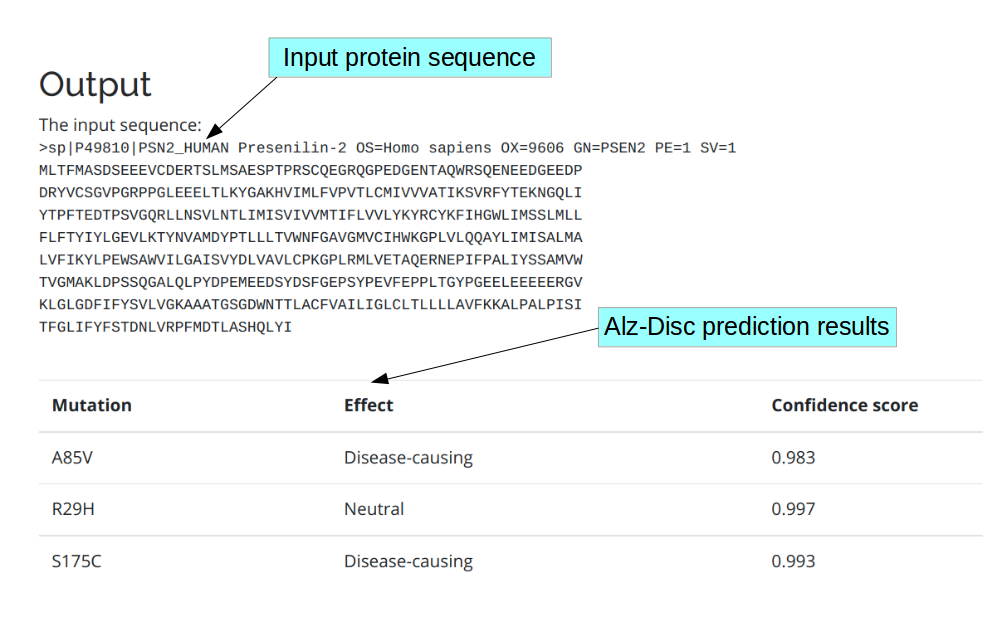
After submitting your job, the protein sequence and followed by prediction results (in the table format) are displayed in the result page. Prediction results includes, mutation information, effect (disease-causing or neutral), confidence score of the prediction.
3. Is Alz-Disc takes the multiple mutations as input?
Yes, you have provide the multiple mutations with comma separated values. Prepare multiple missense mutation using the following format:
Syntax: Mut1{,}Mut2{,}Mut3{,}etc.
Example: A85V,R29H,S175C
4. What is the performance of Alz-Disc? is it better than existing methods?
Alz-Disc method obtained the 91.3%, 95.5%, and 93.5% of sensitivity, specificity and accuracy on 10-fold cross validation, respectively. Besides, it outperformed for the blind test dataset with accuracy of 90%. Yes, Alz-disc showed that improvement of accuracy 13%, which is higher than existing methods available in the literature.