Introduction
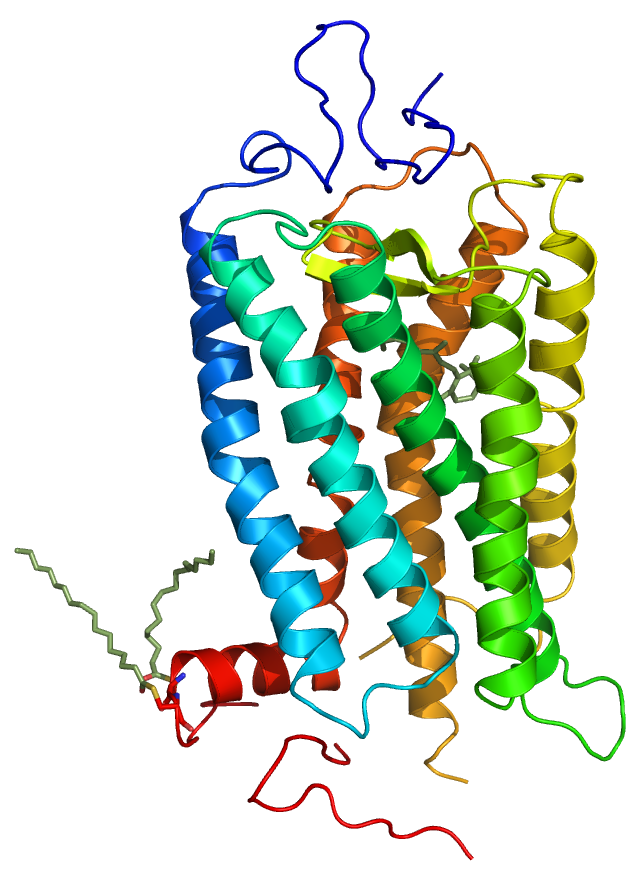
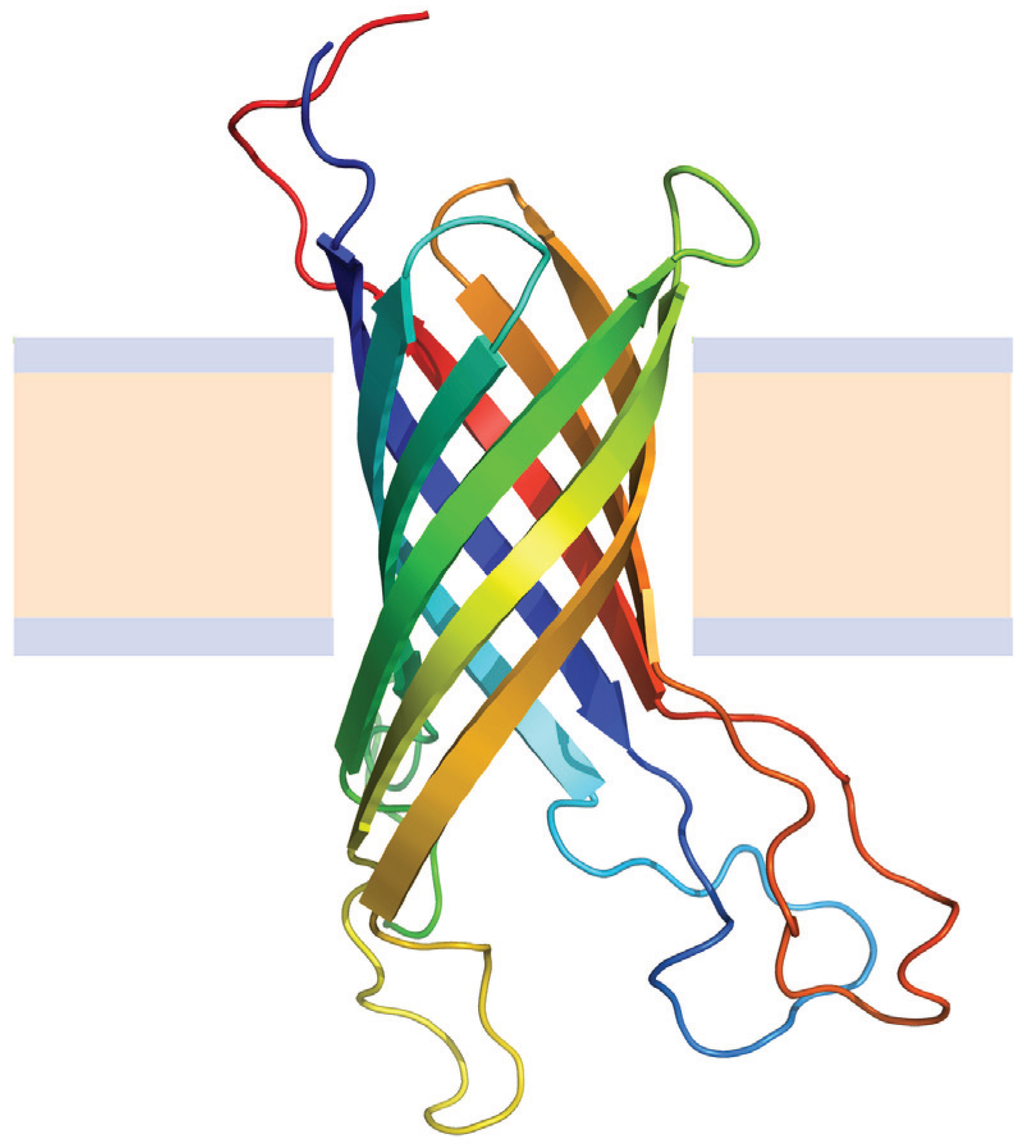
MPTherm-pred is a webserver contains diferent topological specific models for predicting the membrane protein thermal stability upon missense mutations. This method is developed by a set of 929 membrane protein mutations in both membrane bound and aqueous (cytoplasmic/extra-cellular) regions, which are retrieved from MPTherm database. Each model was validated with different cross-validation procedures and tested against evolutionary independent protein mutations. The membrane models showed a pearson correlation of 0.73 with mean absolute error (MAE) of 2.75℃ between experimental and predicted ΔTm values for the test dataset. Similarly, the average correlation and MAE in aqueous regions were 0.75 and 3.5℃, respectively. This suite of predictive methods help in designing the new stable mutants for different applications and to elucidate the key factors influencing the membrane protein stability upon mutations.
Other membrane protein databases and tools
MPTherm
MPTherm is a thermodynamic/stability database of membrane proteins and their mutants.
MutHTP
MutHTP is a comprehensive database of human neutral and disease variants of membrane proteins.
Pred-MutHTP
Pred-MutHTP is a tool for discriminate the disease-causing and neutral mutations of membrane proteins from sequence.
BarodaTM
BorodaTM is a structure based predictor which predicts the effect of the missense mutations in transmembrane proteins.