OVERVIEW
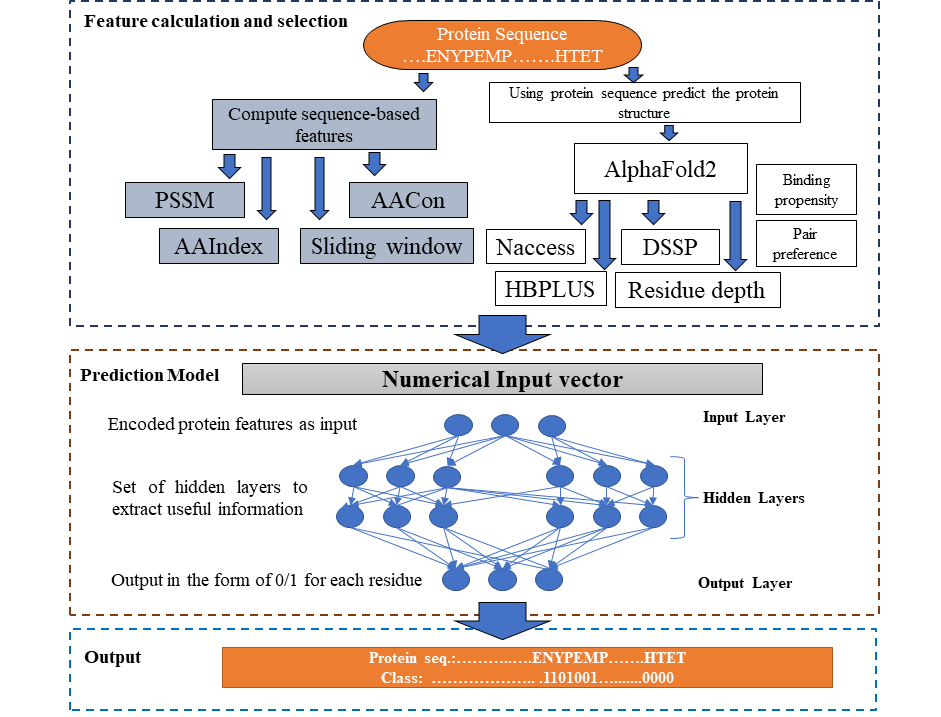
We developed DeepBSRPred, a binding site residue prediction method using protein sequence and predicted structures from AlphaFold2. DeepBSRPred is a deep neural network that uses sequence-based features like position-specific scoring matrix (PSSM), solvent accessibility, conservation score, and amino acid properties from the AIindex database. also, we utilized the structure prediction capabilities of AlphaFold2 and used the predicted structure information to calculate features such as residue depth, and our method shows better performance with an average F1 score 0.73 and MCC of 0.22, which is better than existing methods.