|
PRA-PredProtein-RNA complex Binding Affinity Prediction |
PRA-Pred
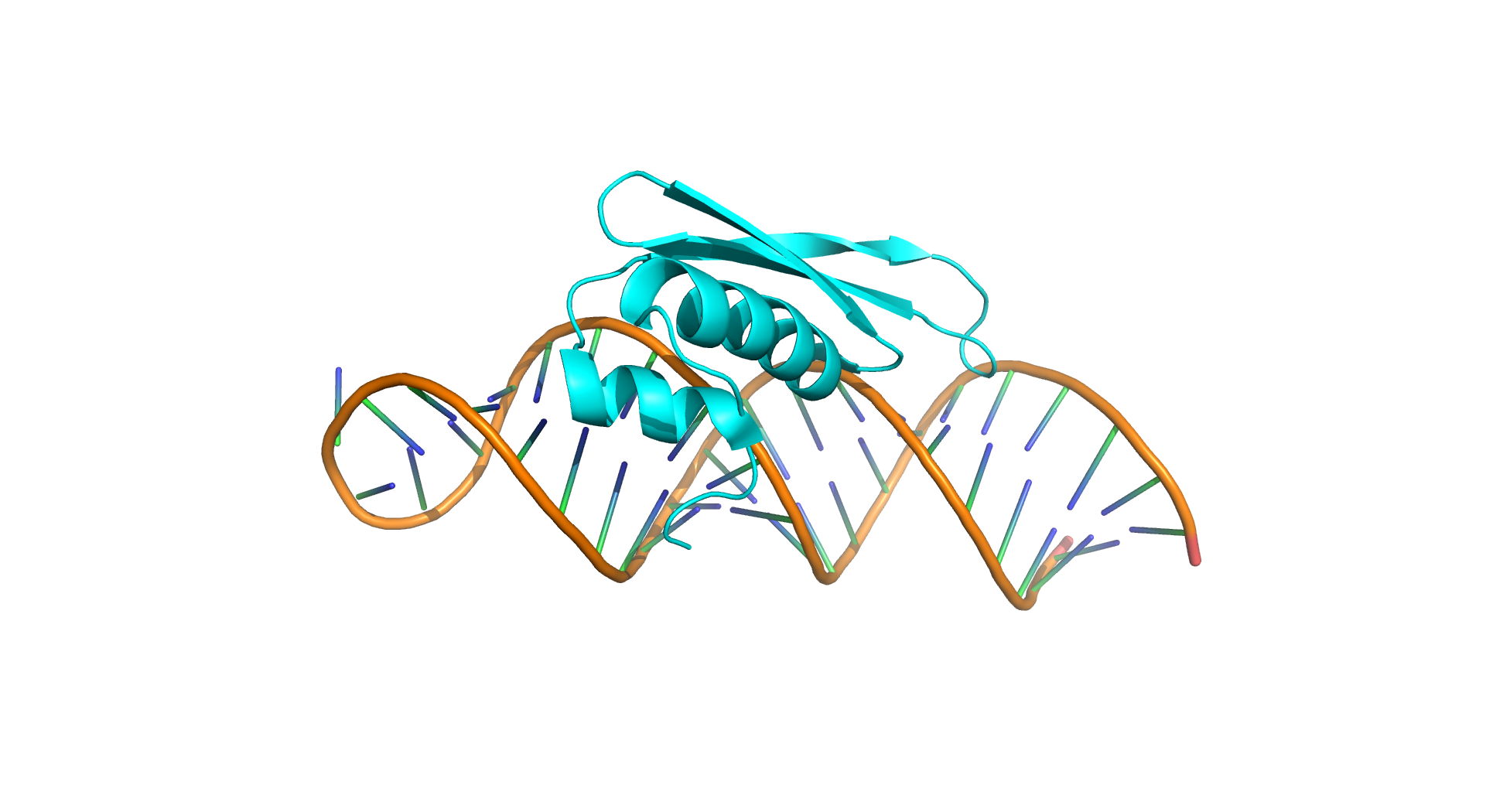
This server predicts protein-RNA binding affinity using structure-based features and classes based on RNA strand, structure and function of protein. We found that RNA base-step parameters, number of atomic contacts between the RNA and protein, interaction energies, contact potentials, and hydrogen bonds are important to understand the binding affinity. PRA-Pred shows a correlation of 0.85 and a mean absolute error(MAE) of 0.81 in Self-consistency, correlation of 0.77 and a MAE of 1.02 kcal/mol in jack-knife test.
Please click here to start prediction.