Using ProAffiMuSeq
Prepare your input
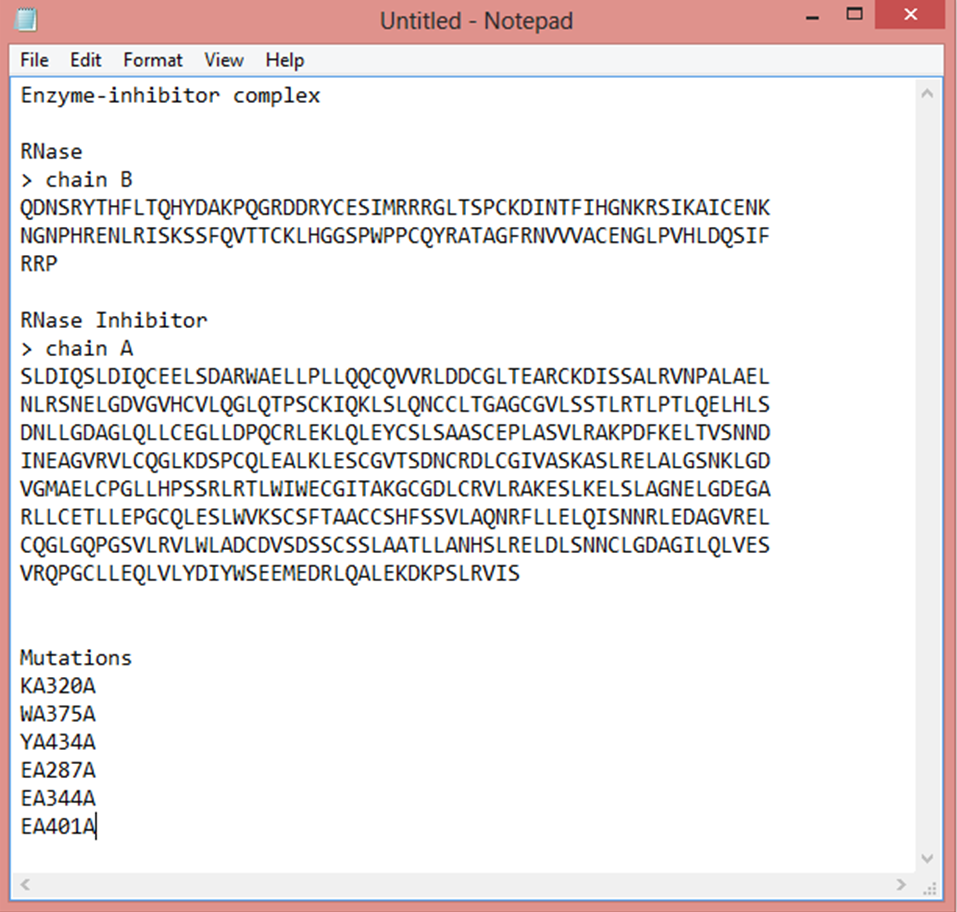
Keep the sequences and mutations and functional information (including the function for each protein partner). For example, in an enzyme-inhibitor complex, the binding partners are either enzymes or inhibitor proteins.
Input functional information and sequences
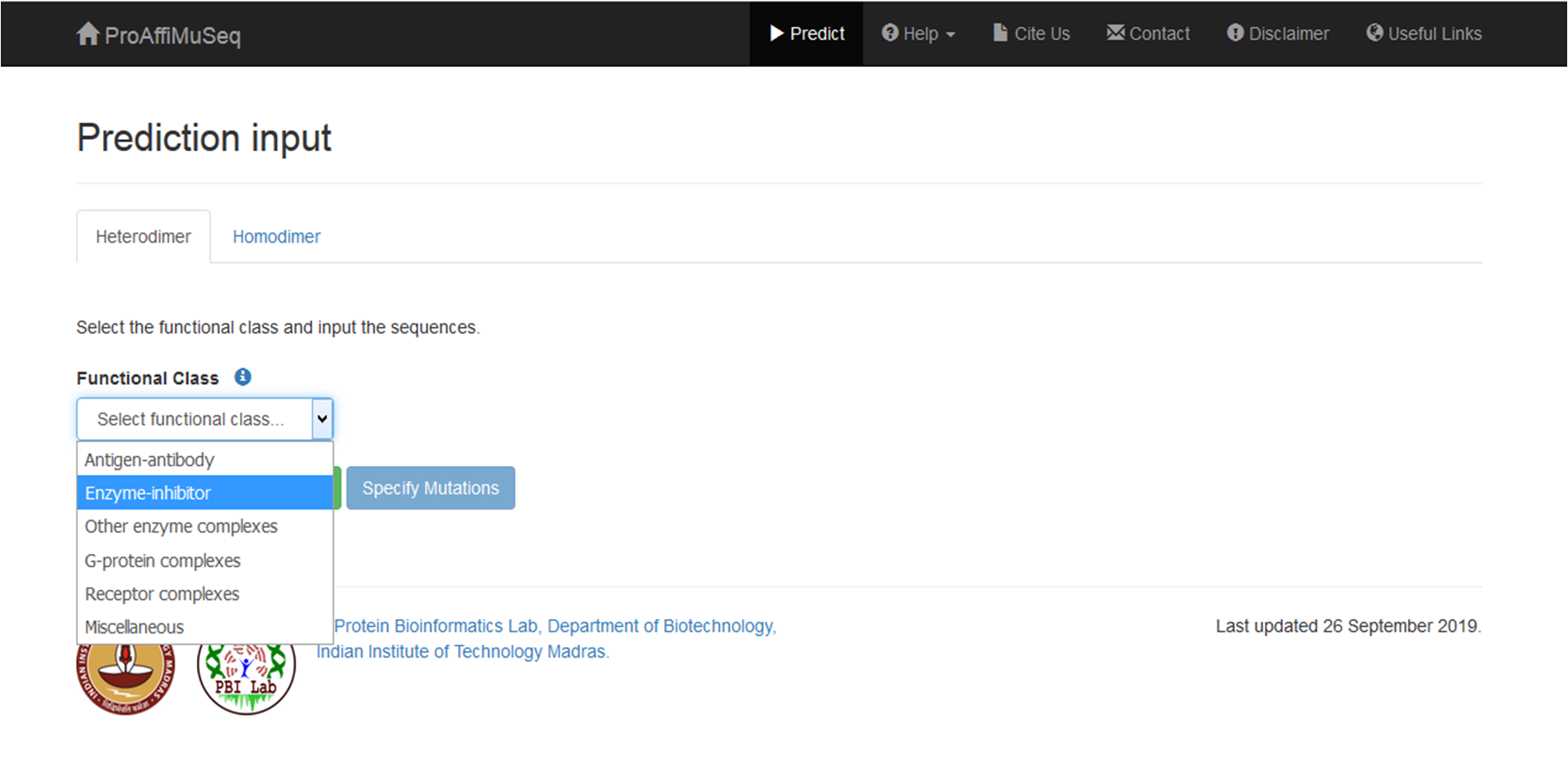
Select the functional class of the protein-protein complex. If the protein-protein complex does not fall under the given categories, choose 'Miscellaneous'. We recommend knowing the functional class as this will result in better accuracy and the predicted ΔΔG value is more likely to reflect the actual change in binding affinity.
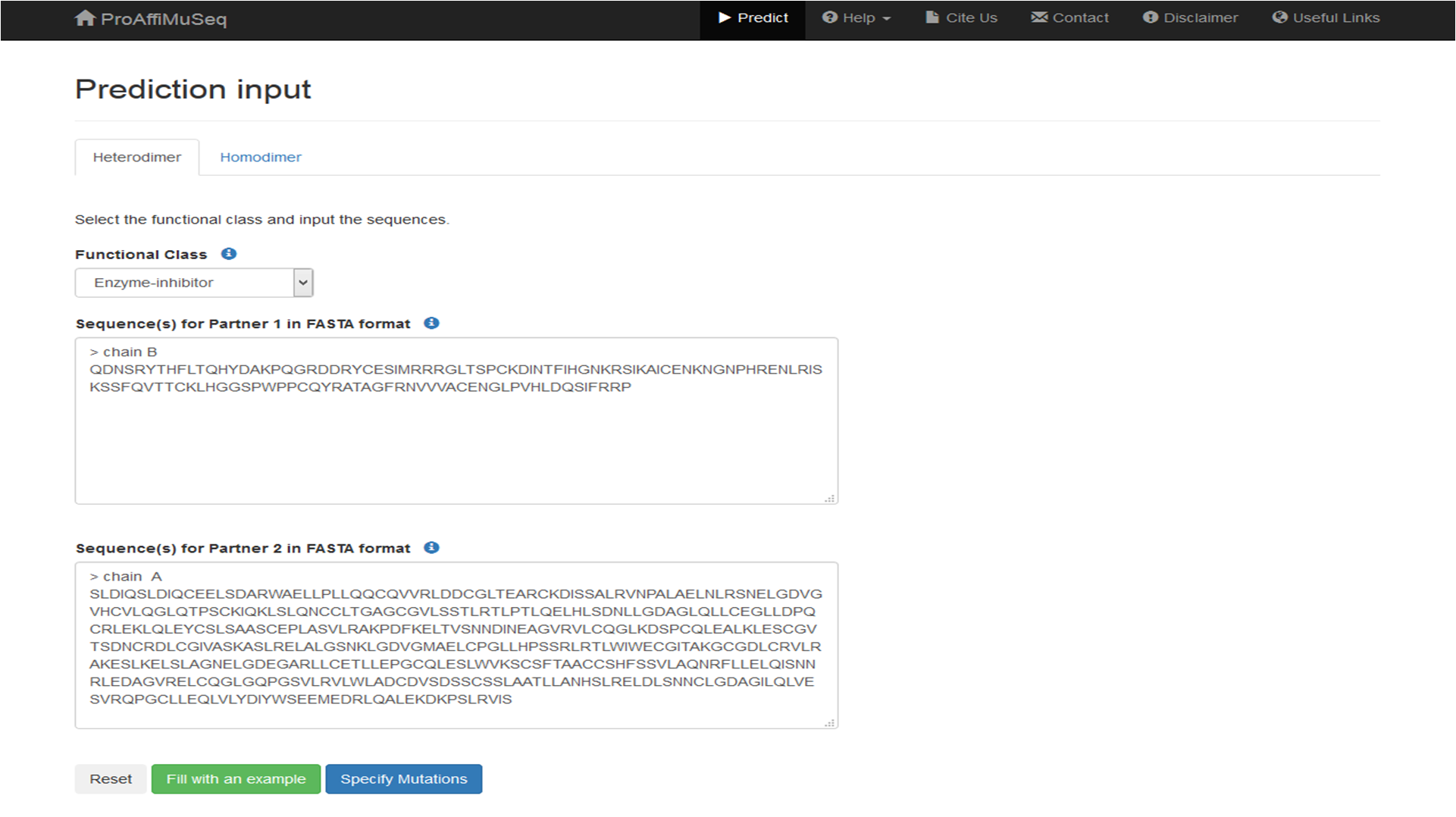
Textboxes will appear for sequence inputs, based on the functional class selected. Input the sequences accordingly. For example, for an enzyme-inhibitor complex, input the enzyme sequence in the first box and the inhibitor sequence in the second box.
Click on 'Specify Mutations'.
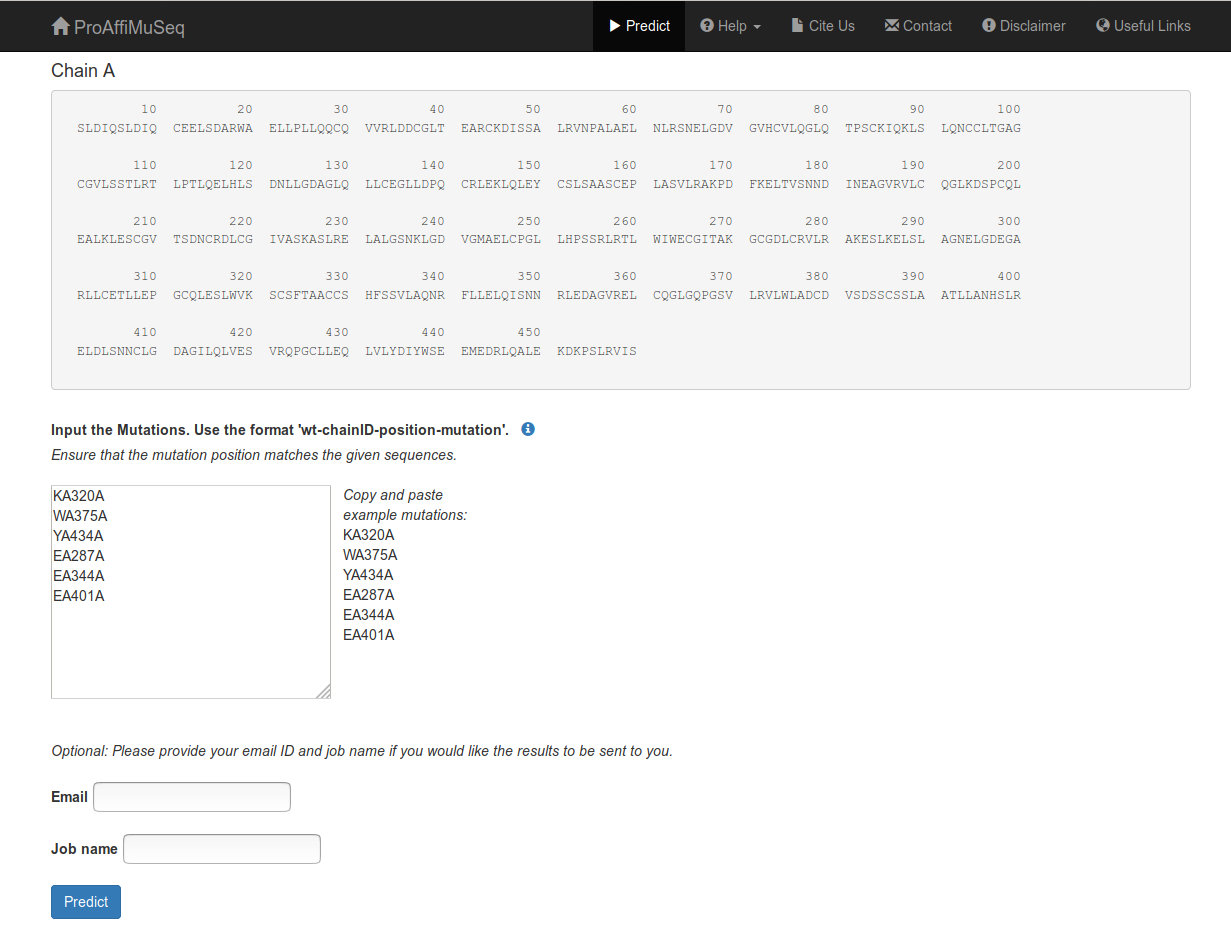
Input mutations
Provide all mutations in the specified format: 'wt-chainID-position-mutation'. Input one mutation per line only. Check with the sequences to ensure that the wild-type residue matches with the sequence at the given position.
You can also provide an email address and a job name, if you wish to get the results by email.
Click on 'Predict' to run the job.
While the server processes your job...
The server usually takes 1 minute per sequence to calculate the features and predict the results. Please be patient and DO NOT CLOSE the browser window.
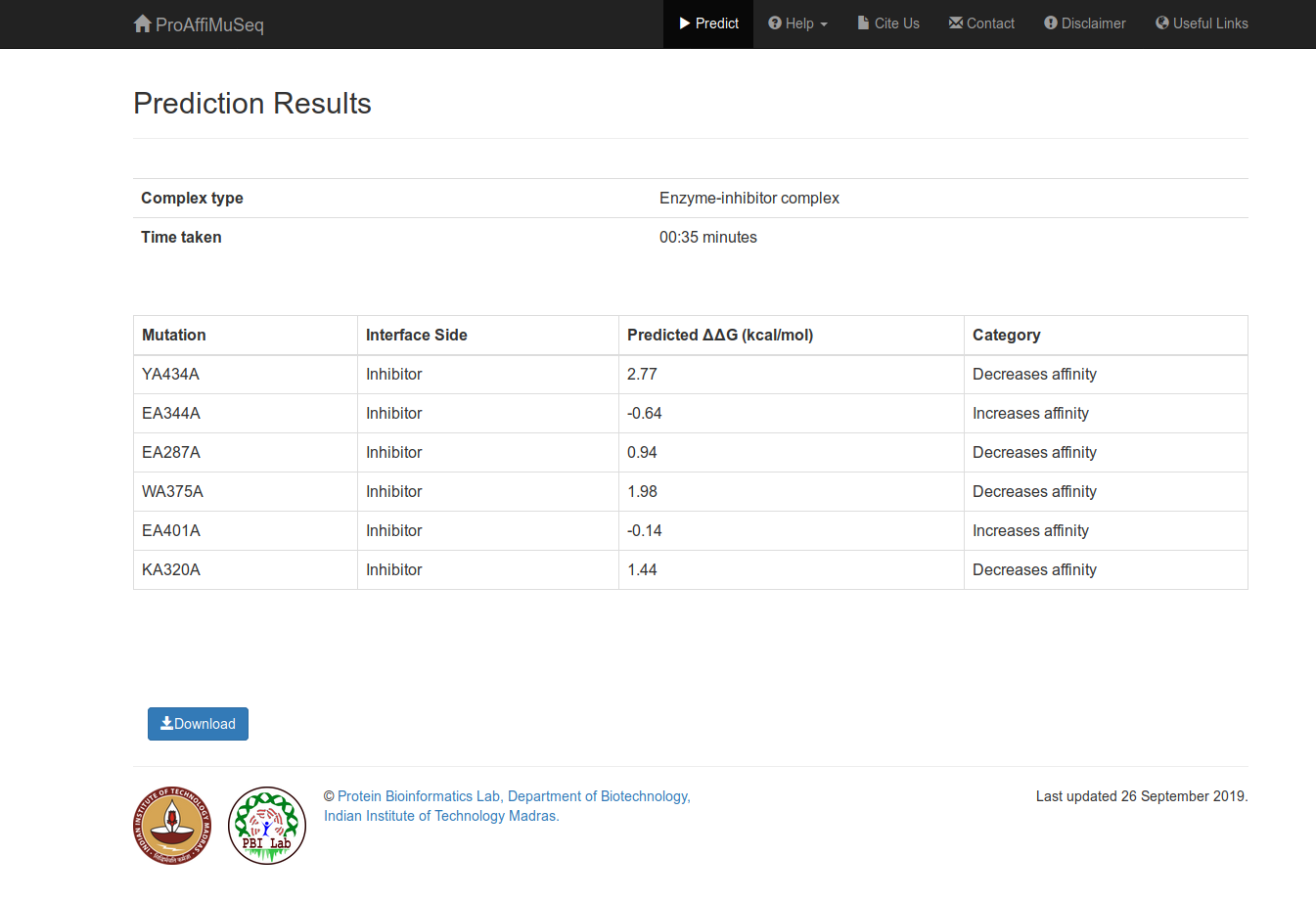
Results
The results will be displayed in tabular format. You can also download the results as a text file.