VEPAD accepts first 5 columns of a VCF file without header as input. Five of the columns being chromosome number, position on the chrosome, label, reference allele and the alternate allele. The same can be downloaded here.
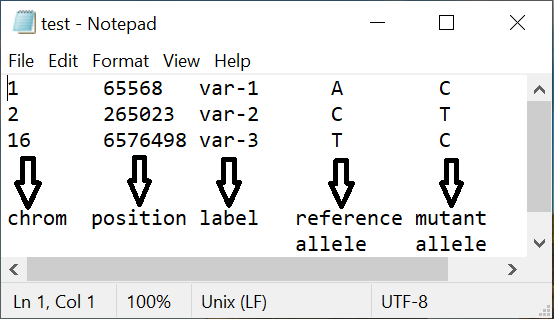
2. What is the output of VEPAD?
The result contains the score and the remark (benign or deleterious) assigned by the tool to each variant submitted by the user. This score signifies the probability of a variant being deleterious in context with Alzheimer's Disease.
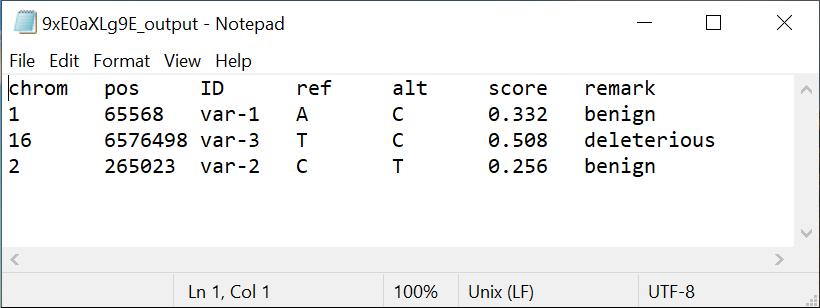
3. How to interpret the scores assigned to the mutations by VEPAD?
We categorise mutations into 2 categories based on the score assigned by our model. Mutations with a score of less than 0.38 are labeled as "benign" and mutations whose score is greater than 0.38 are labeled as "deleterious".
4. How to use VEPAD for large number of mutations?
Kindly contact us at vepad.iitm(at)gmail.com to obtain the standalone version of VEPAD.