ANuPP - Tutorial
Aggregation Nucleation Prediction in Peptides and Proteins (ANuPP) is a classifier-classifier developed and trained to identify amyloid-fibril forming peptides and regions in protein sequences. Following tutorial explains some of the terms used in this site. To know more about the methodology, please read the article (Ref.). The ANuPP prediction can used to
- 1 Prediction of ammyloidogenic hexapeptides
- 2 Identify potential APRs in the Sequence
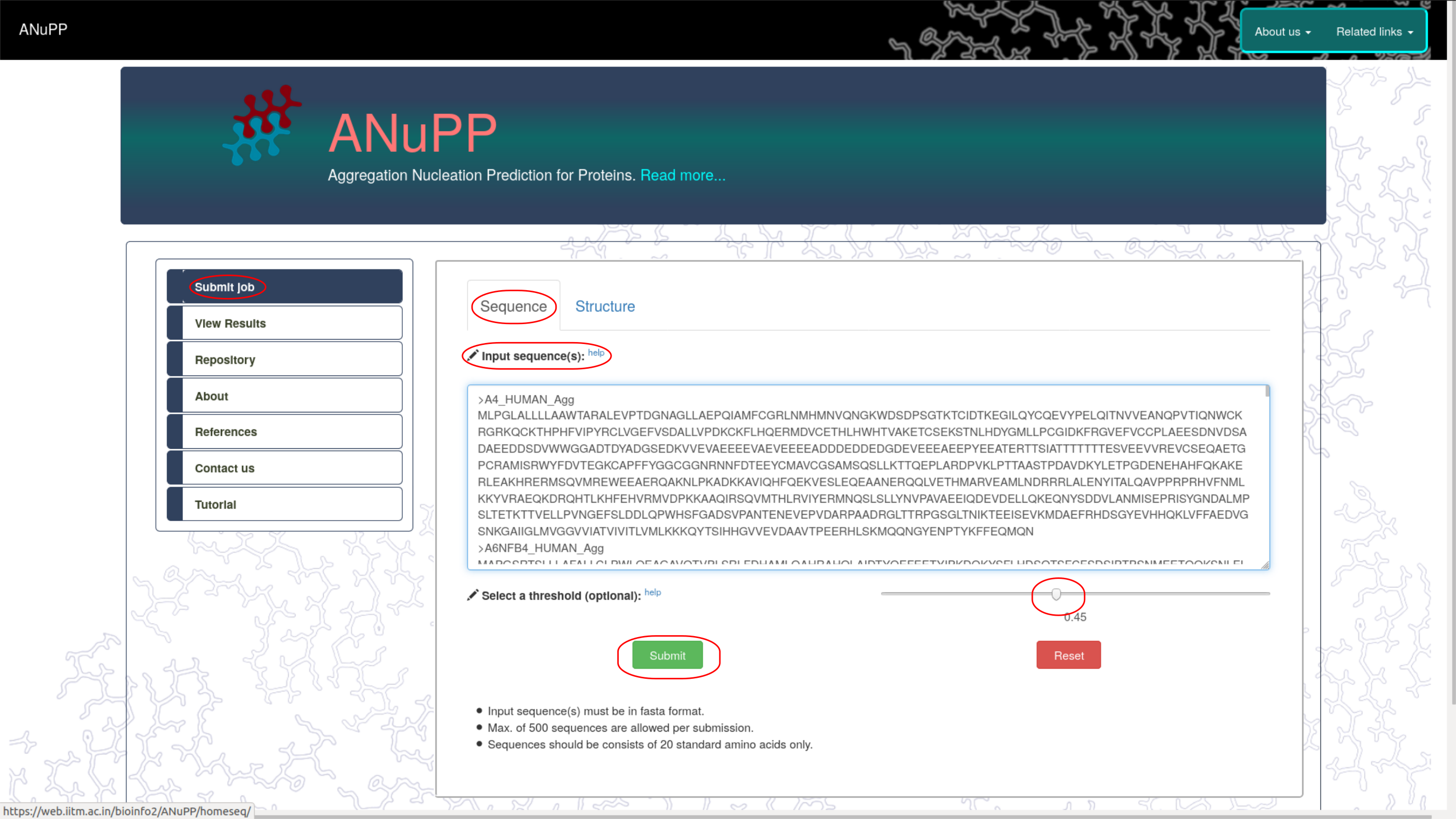
- ANuPP is a sequence-based predictor.
- ANuPP can read the sequence as fasta format or extract from the input PDB structure.
- ANuPP accepts 50,000 sequences in a single run.
- Users can adjust the probability threshold cut-off to increase or decrease senstivity/specificity.
- On submission, the sequences/structure is processed by ANuPP and the results are listed within a minute.
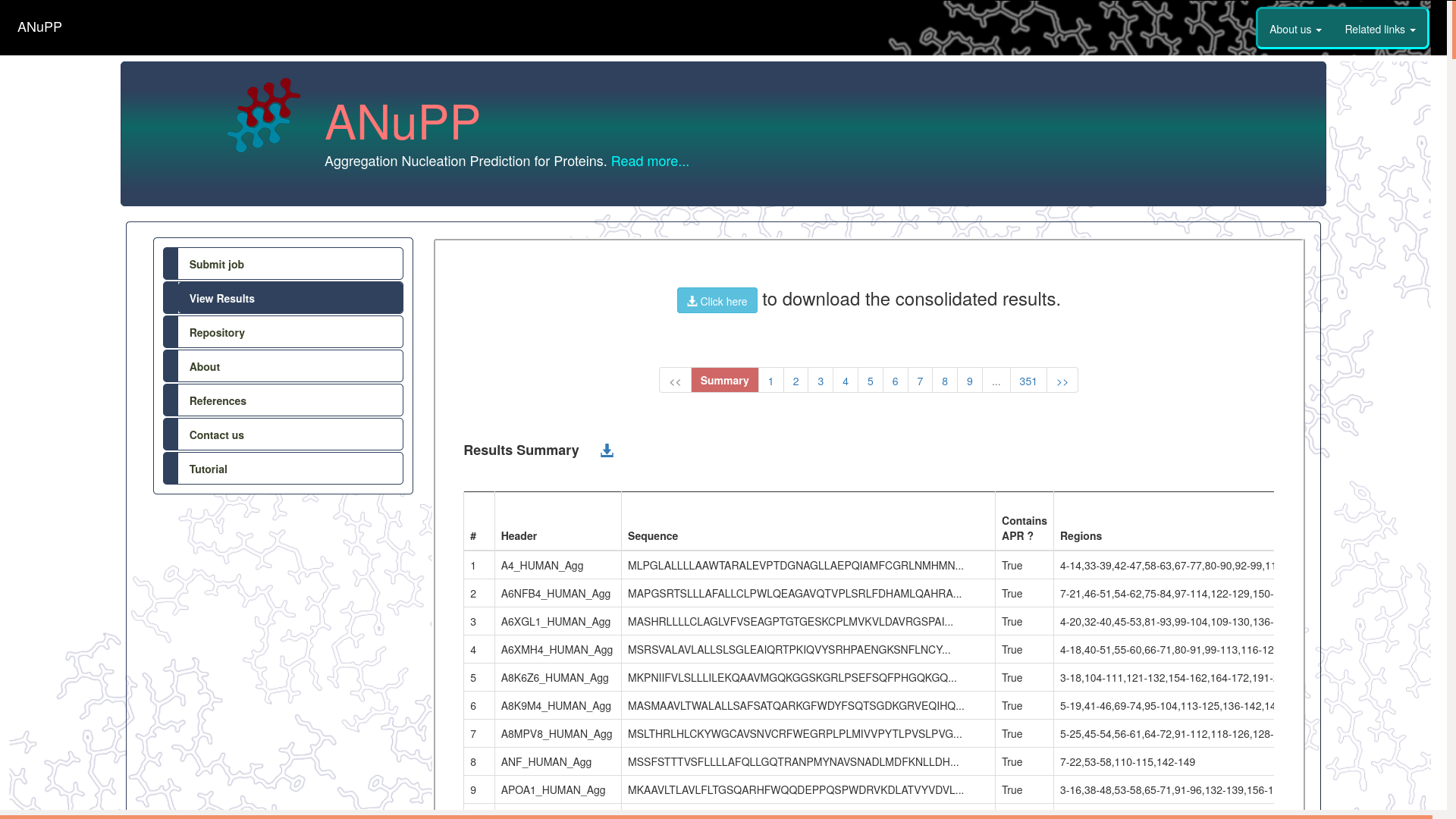
- The results are displayed as a consolidated summary and a series of detailed results page, one for each submitted sequence.
- The summary page show below provides wide-range of statistics on the predicted ARPs and the aggregation propensity.
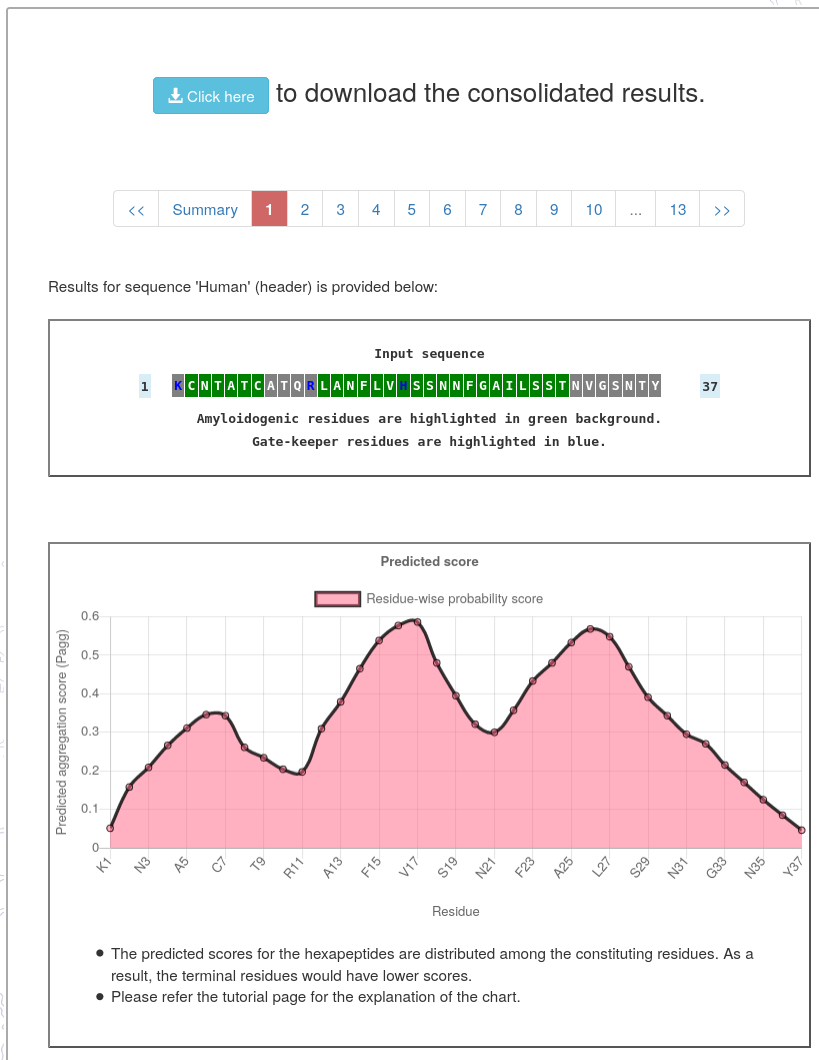
- The detailed result page provides predicted aggregation score for the residues and hexapeptides in the given sequence.
In addition, ANuPP server also acts as a repository for the ANuPP prediction results. The database section of the server contains two subsections:
- 3 Human proteome
- 4 Hexapeptides
The "Human proteome" subsection lists the predicted APRs in human proteins along with relevent details. The "Hexapeptides" subsection lists the prediction score of all 64 million peptides. We believe the data presented in the database section could be useful to the scientifc community. In addition to ANuPP predictions, we have included predictions from TANGO and WALTZ scores for 64 million hexapeptides.
Incase of any requests or queries, please contact us.