Overview
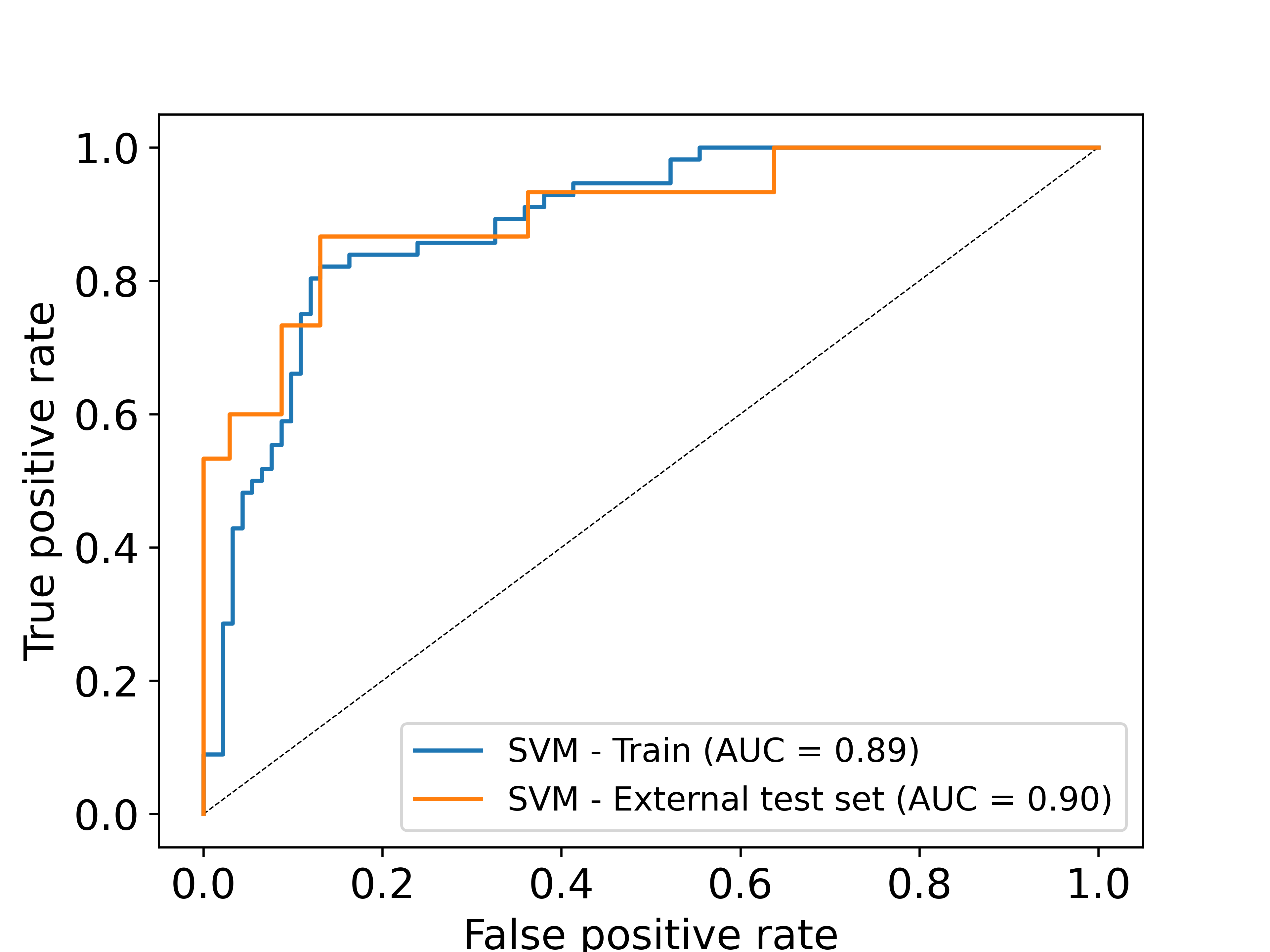
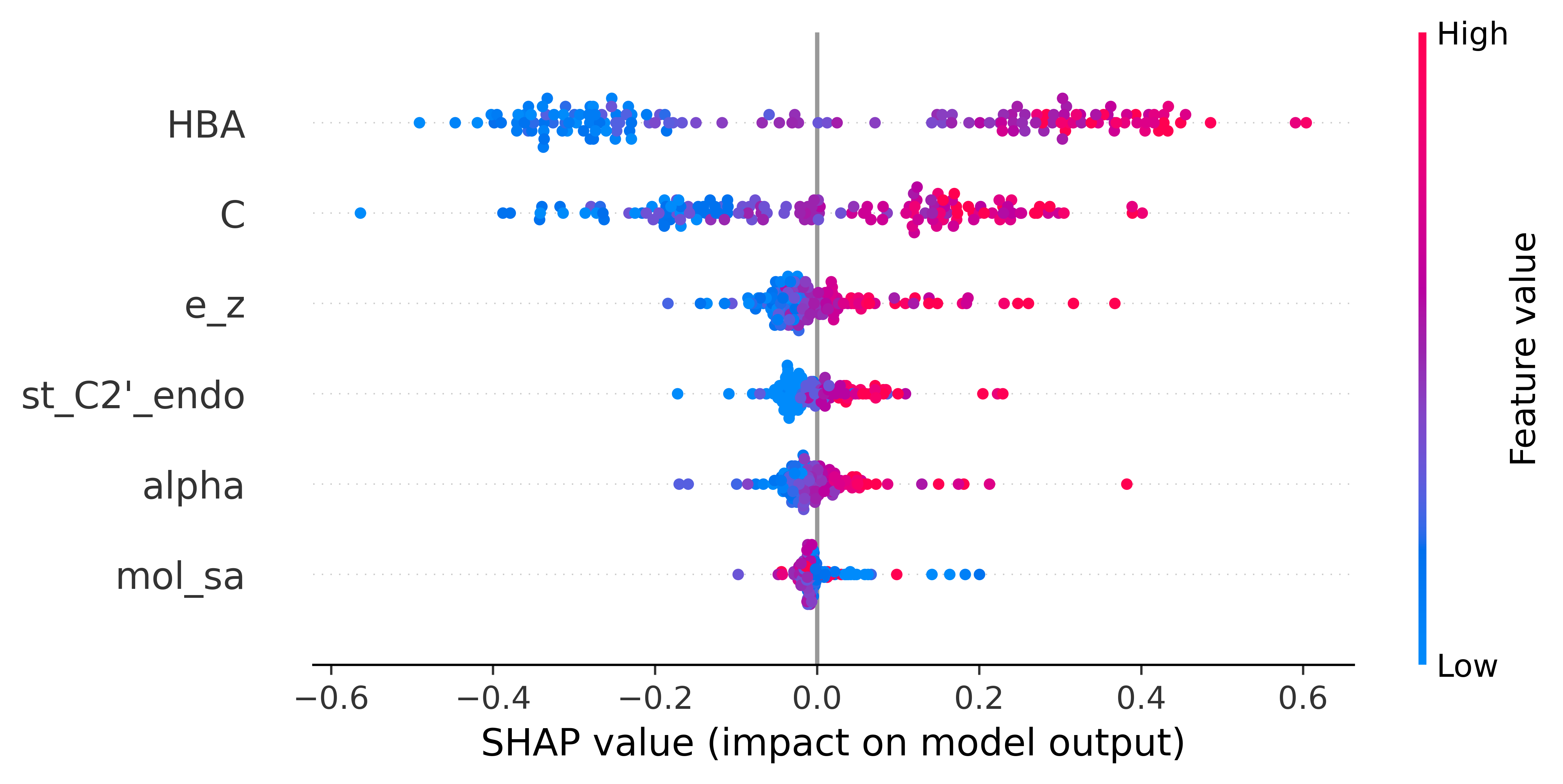
DRLiPS is a method to predict druggable small molecule binding sites in 3D structures of RNA targets, using a combination of RNA structure-based features. DRLiPS provides a ranking of all possible binding sites in a given RNA target, in terms of their ability to selectively interact with a drug-like small molecule. The druggability scores per binding site provided by DRLiPS are the probability values from the underlying machine learning model powering this method. Upon evaluation on an external blind test dataset of RNA targets, DRLiPS perfomed with an F1 score of 0.80 and ROC of 0.81, which are the current state-of-the-art metrics compared to existing methods in this field. For more details on the datasets and model performance, please visit our statistics page.
DRLiPS can accept a PDB file of the RNA target along with a list of binding site residues lining the cavity of interest. If a binding site is not provided, DRLiPS proceeds to predict all possible binding sites in the structure using a standard prediction program, and rank them based on their druggability score. Users can check our tutorial page for further details on using the web server and interpretation of results. If you have any further queries regarding the server or the models, please visit the FAQs page or write to us. Thank you for using DRLiPS!