Simple search
Under simple search, the user can search based on either the Entry ID or a Keyword.
Search by Entry ID
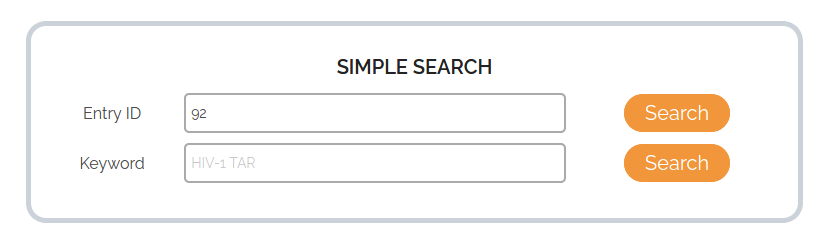
To search by Entry ID, the user can enter a number from the available entries (1 to 2050). In the example above, we are searching for the database entry defined by the Entry ID 92. The results of Entry 92 will be displayed as shown below.

Search by Keyword
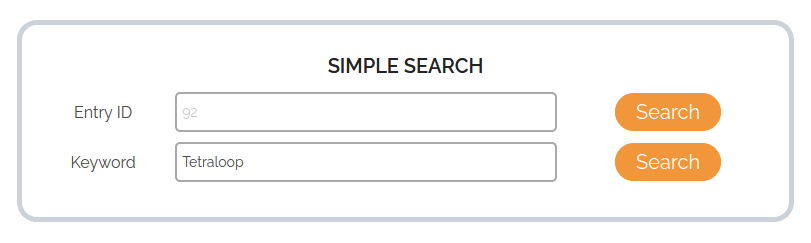
To search by Keyword, the user can enter any word describing the RNA target name, small molecule name, RNA sub-type, RNA class, RNA secondary structure, experimental method or reference-related information such as keywords from title, PubMed ID and authors involved. In the example above, we are searching for RNA-small molecule interactions involving RNA targets with a Tetraloop secondary structure. The results from the search will be displayed as shown below.

The search results are tabulated with the default display options for search (Users can modify the display options using Advanced search). The number of search results matching the input keyword is shown above the results table. In the example case, 4 entries were retrieved with the keyword Tetraloop from the database. Users can also sort the results based on any of the table columns by clicking the name of the column in the results table. Upon clicking the Entry ID in the table shown, detailed information corresponding to the entry will be displayed in a new tab. The keyword search option can currently work with only 1 keyword per search. Search with multiple keywords or logical operators is not available.
Advanced search
Advanced search can be used to control both the database search and display options. Users have to select at least one option from the search options available in the left hand panel of the search form. The results from Advanced search will be displayed in a table format identical to the one used in Simple search by Keyword.
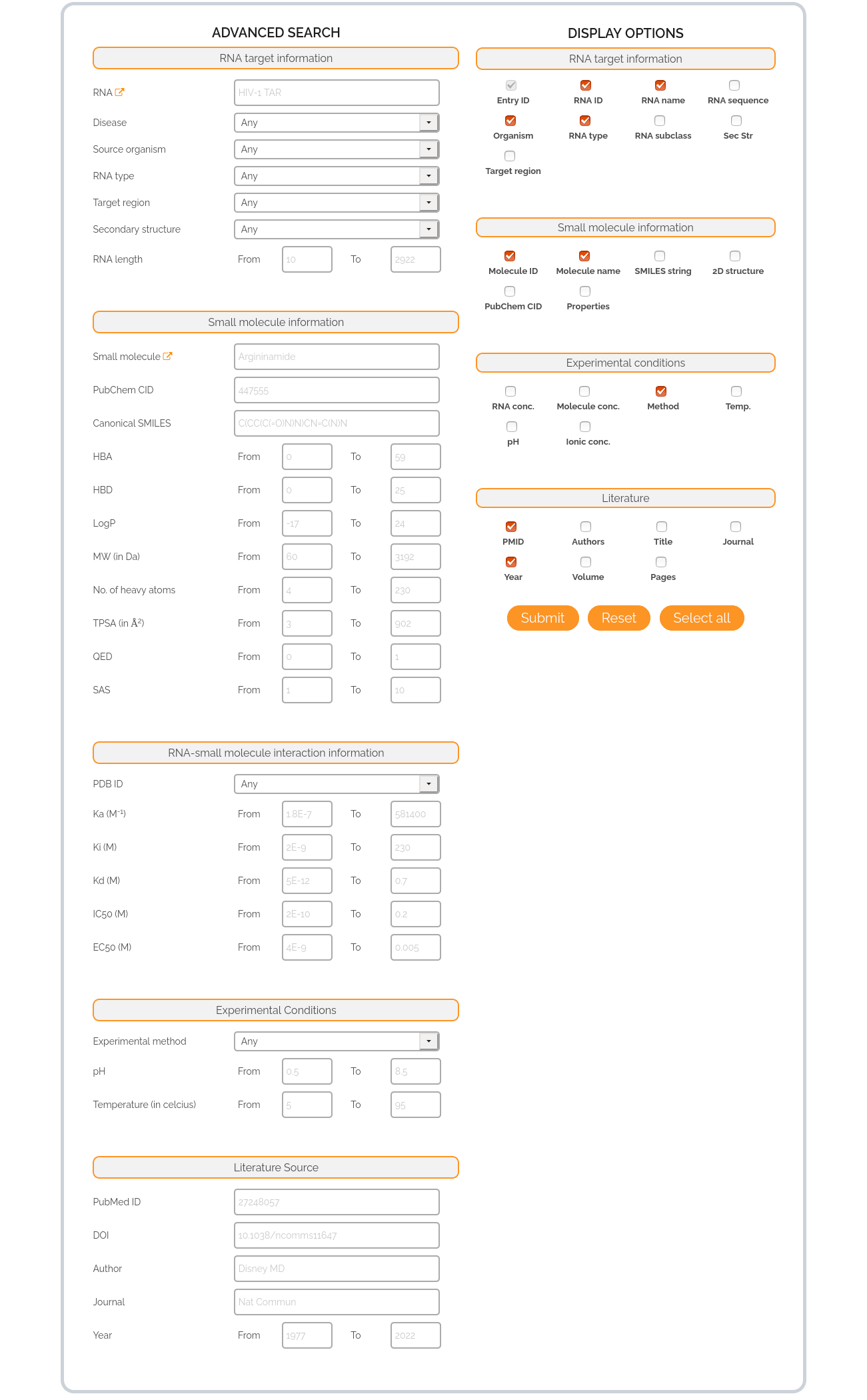
Search Options
RNA target information: This section includes options to search the database based on RNA target name, disease condition, source organism, RNA type, target regions within known RNA targets, secondary structure and the length of the RNA target. Except the RNA target name and RNA length, other options are displayed in dropdowns where, the available RNA types, disease conditions, organisms, target regions and secondary structures are updated based on the entries available in the database. For the available list of RNA targets in R-SIM, a hyperlink is provided beside the RNA option.
Small molecule information: This section includes options to search the database based on small molecule name, PubChem CID (wherever available), canonical SMILES and filtration criteria based on the key physicochemical properties of molecules (HBA, HBD, LogP, MW, No. of heavy atoms, TPSA, QED and SAS). Users can curate virtual screening datasets of small molecules in any required property space, by application of these filters. For a brief description of each molecular property, kindly check the Glossary tab under Tutorials. For the available list of small molecules in R-SIM, a hyperlink is provided beside the Small molecule option.
RNA-small molecule interaction information: This section includes options to search the database based on PDB ID of an existing RNA-small molecule complex or based on different ranges of affinity exhibited by small molecules to RNA targets. In case of RNA-small molecule complexes whose structure is not known, the user can search based on the other available options specific to RNA and small molecules.
Experimental Conditions: This section includes options to search the database for entries validated by a specific experimental method. The list of available experimental methods in the R-SIM database is provided in the dropdown from which users can choose only one option at a time. Further, options to choose a specific range of pH and temperature are also available.
Literature Source: This section includes options to search the database for entries from specific references or articles from literature. Users can search for RNA-small molecule interactions reported in a published article based on: PubMed ID, DOI, author(s), journal name and year of publication. Users can also supply a range for the publication year, to retrieve all RNA-small molecule interactions reported in literature published in that year range.
Display Options
The default display options are shown as checked boxes in the form. Users can customize the display options by modifying the check boxes according to their need, after selecting at least one search option in the left hand panel of the advanced search form.
CASE STUDIES
-
How to retrieve data on RNA targets involved in Cancers?
-
How to retrieve data on small molecules with LogP in range [0,3], QED in range [0.3,0.4] and SAS in range [1,3]?
-
How to retrieve all RNA-small molecule interactions published from 2000 to 2002 in R-SIM?

Step 1: From the options listed under RNA target information, select Disease dropdown and choose the Cancer option.
Step 2: Select the necessary display options from the right hand panel to customize the search results page.
Step 3: The search results will be displayed in a tabulated form containing results specific to the query parameter(s). In this example, the results table should contain 342 entries involving RNA targets attributed to various Cancer types.
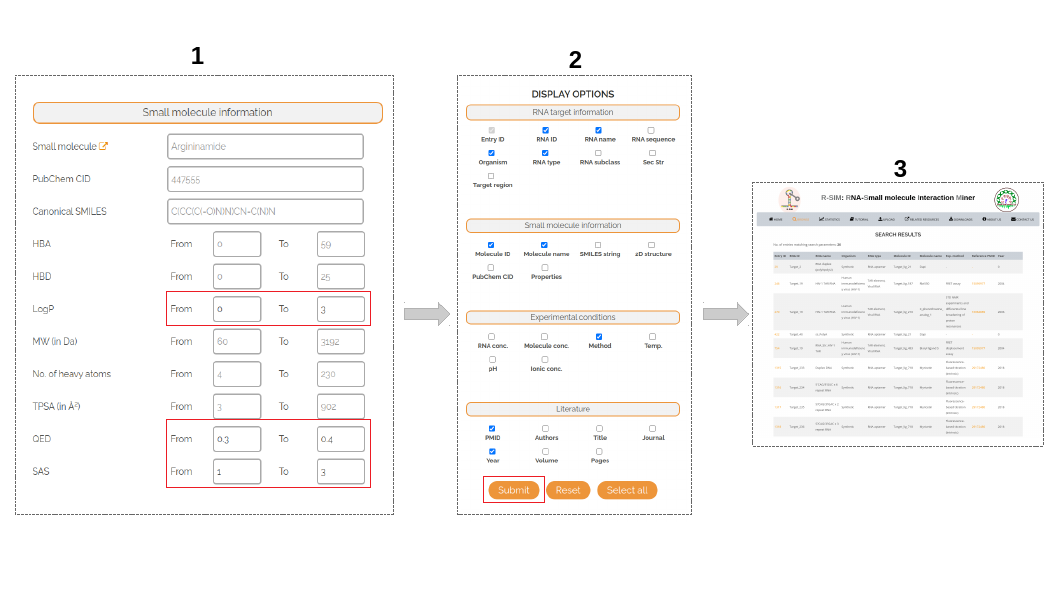
Step 1: From the options listed under Small molecule information, update the From and To ranges for LogP, QED and SAS properties to the desired values.
Step 2: Select the necessary display options from the right hand panel to customize the search results page.
Step 3: The search results will be displayed in a tabulated form containing results specific to the query parameter(s). In this example, the results table should contain 26 entries involving small molecules satisfying the desired property ranges for all 3 properties of interest.

Step 1: From the options listed under Literature Information, update the From and To ranges for Year to the desired values.
Step 2: Select the necessary display options from the right hand panel to customize the search results page.
Step 3: The search results will be displayed in a tabulated form containing results specific to the query parameter(s). In this example, the results table should contain 57 entries involving RNA-small molecule interactions published between 2000 and 2002 available in R-SIM.
DATA UPLOAD GUIDELINES
The Data Upload form has been designed to allow users to contribute to R-SIM by submitting experimentally verified RNA-small molecule interactions from already published articles or from their experimental studies. Before uploading data to the R-SIM database, users are requested to go through the data upload guidelines provided below to ensure that their data satisfies our evaluation and becomes part of R-SIM during successive updates. Thank you for sharing your data with us!
Please ensure that the contact information (specifically the e-mail ID) is correct. This is essential on submission of experimental information to ensure correctness of data in the database. All personal details entered in R-SIM will be maintained confidential and will not be shared to third parties without consent.
Please ensure that the data entered in the upload form is published, experimentally verified and has been indexed with either a PubMed ID or DOI. Data supplemented with an invalid DOI or PubMed ID will not be included in R-SIM.
Even though PDB ID field is not mandatory, the users are strongly recommended to provide PDB ID wherever the RNA-small molecule complex structure is available.
When providing the RNA sequence information, kindly include the source of the sequence within paranthesis. Source can be any article DOI or accession identifiers from established sequence databases.
When providing the SMILES string for the small molecule, please ensure that the SMILES has been canonicalized, includes stereochemistry information and is valid (for further use with RDKit or OpenBabel packages).
Kindly ensure that the temperature is in Celcius units and the binding affinity value is in Molar units only. For all affinity values, please use scientific notation ('1E-9').
All data uploads are subject to verification and redundancy checks prior to inclusion in R-SIM.
Glossary of terms
| Terms | Explanations |
|---|---|
| Molecule_name | Name of the small molecule |
| Molecule_ID | Entry number of the small molecule in the database. This number can be used to get all information relevant to the molecule during search. |
| Target_RNA_name | Name of the RNA target |
| Target_RNA_ID | Entry number of the RNA target in the database. This number can be used to get all information relevant to the RNA during search. |
| Ka(M) | Association constant of the small molecule in molar units |
| Ki(M) | Inhibition constant of the small molecule in molar units |
| Kd(M) | Dissociation constant of the small molecule in molar units |
| IC50(M) | Half-maximal inhibitory concentration of the small molecule in molar units |
| EC50(M) | Half-maximal effective concentration of the small molecule in molar units |
| T50(M) | Half-maximal activation constant of the small molecule in molar units |
| PDB_ID | Protein Data Bank code for the RNA-small molecule complex, wherever available |
| Modeled_complex | Link to the modeled RNA-small molecule complex obtained using RDock program |
| Reference_DOI | The Digital Object Identifier (DOI) of the reference article or the source article for the RNA-small molecule interaction |
| Reference_PMID | Link to the PubMed identifier of the reference article |
| Title | Title of the reference article |
| Authors_list | List of authors for the reference article |
| Journal_name | Name of the journal where the reference article has been published |
| Volume | Volume of the journal in which the reference article has been published |
| Page_number | Page numbers in the specific volume of the journal where the reference article has been published |
| Year | Year in which the reference article was published |
| Ligand_concentration | Concentration of the small molecule (ligand) used for experimental validation of the RNA-small molecule interaction. Units of the quantity are retained as they appear in the reference article. |
| RNA_concentration | Concentration of the RNA target used for experimental validation of the RNA-small molecule interaction. Units of the quantity are retained as they appear in the reference article. |
| pH | pH at which the experimental validation was performed |
| Temperature | Temperature at which the experimental validation was performed |
| Ions_present | Names and concentrations of the various ions (as ionic salts or buffers) used in the experiment |
| Experimental_method | Name of the experimental method used to verify the RNA-small molecule interaction |
| Data_source | Source database from which the interaction was taken. For data that are not part of any previously known databases, "Additional" tag is used here. |
| SMILES | Simplified Molecular Input Line Entry System (SMILES) identifier for the small molecule |
| IUPAC_name | The standard IUPAC name for the small molecule, wherever available |
| Pubchem_CID | Link to the PubChem chemical identifier page of the small molecule, wherever available |
| LogP | Predicted octanol-water partition coefficient of the small molecule (from RDKit) |
| MW | Molecular weight (in Da) of the small molecule (from RDKit) |
| HBA | Count of hydrogen bond acceptors in the small molecule (from RDKit) |
| HBD | Count of hydrogen bond donors in the small molecule (from RDKit) |
| NRB | Number of rotatable bonds present in the small molecule (from RDKit) |
| TPSA | Topological Polar Surface Area (in Å2) of the small molecule (from RDKit) |
| Benzene_count | Number of benzene rings present in the small molecule (from RDKit) |
| QED | Quantitative estimate of drug-likeness of the small molecule (from RDKit) |
| SAS | Synthetic Accessibility Score of the small molecule (from RDKit) |
| Heavy_atom_count | Number of heavy atoms (non-hydrogen atoms) present in the small molecule (from RDKit) |
| Target_RNA_sequence | Primary sequence of bases present in the RNA target |
| Sequence_source | Source of the RNA primary sequence information |
| RNA_type | Type of RNA target (mRNA, miRNA, lncRNA, viral RNA etc.,) |
| RNA_subclass | Subclass of RNA target (pre-miRNA, pri-miRNA, FSS element, TAR element, Riboswitch etc.,) |
| RNA_sequence_length | Length of the RNA primary sequence (in bases) |
| Target_region | Region where the binding site for the small molecule can be found (5'-UTR, 3'-UTR, Splice site etc.,) |
| Secondary_structure | Secondary structure formed by the target region (Stem-loop, Bulge, G-quadruplex etc.,) |
|
Copyright © 2022 - All Rights Reserved. The database is maintained by Protein Bioinformatics Lab Department of Biotechnology, Indian Institute of Technology Madras |
 |