Tutorial
The predict page can be accessed from here. This page takes input of protein sequence, mutations and predicts the change in binding affinity upon mutation.
Input options:
- The page has option to select one of following five classification model for the prediction. *
- Monomer-Monosaccharide
- Oligomer-Monosaccharide
- Protein-Disaccharide
- Protein-Trisaccharide
- Protein-Oligosaccharide
- The page has option to enter protein sequence in fasta format. *
- The page has option to enter UniProt ID (to obtain modelled AlphaFold structure) or upload structure file in pdb format.*
- User has to enter the mutation list. Input one mutation per line only. Check with the sequences to ensure that the wild-type residue matches with the sequence at the given position. *
SERVER INPUT PAGE
The below image shows the input page of the PCA-MutPred.
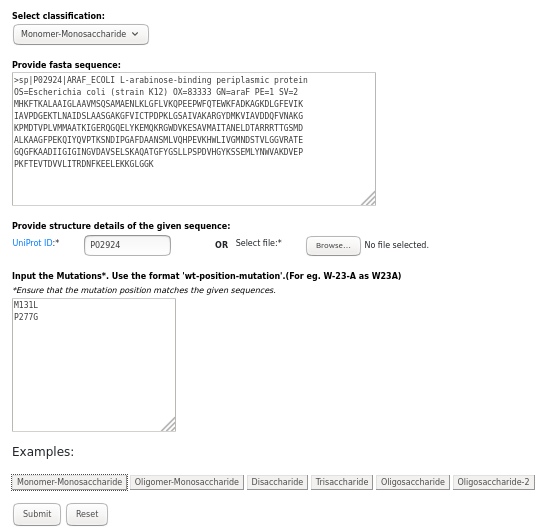
Example:
L-arabinose-binding periplasmic protein (Uniprot ID: P02924)
INPUT:
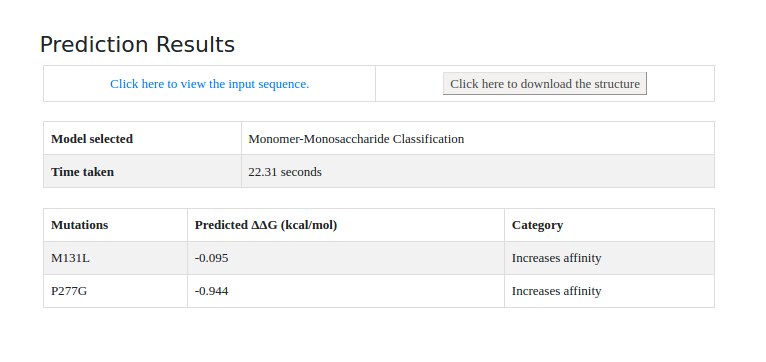
UniProt ID: P02924, belongs to 'Monomer-Monosaccharide' classification and mutation used are M131L and P277G.
RESULT FOR EXAMPLE:
The results will be displayed in tabular format.