|
PDA-PredProtein-DNA complex Binding Affinity Prediction |
 |
Tutorial
The predict page can be accessed from here.
This page takes input of protein-DNA complex and predicts the binding affinity.
Input options:
- The page has option to enter PDB ID of protein-DNA complex or user can upload their file in PDB format. *
- User has option to enter the chain ID of protein. Multiple chain Ids can be entered with comma. (Eg. Chain ID: A,B) (Optional)
- User can enter the chain ID of DNA. Multiple chain Ids can be entered with comma. (Optional)
- User should select one of the DNA class for the prediction. *
- single strand
- Double strand
- User should select one of the strucutral class of protein for the prediction, if the DNA is double stranded *
- all-α
- all-β
- αβ
- others
- User should select one of the Functional class of protein for the prediction. *
- Regulatory
- other (non-regulatory)
- Any problem with the server: Users can download source code from: https://github.com/Harinikannan98/PDA-PreD
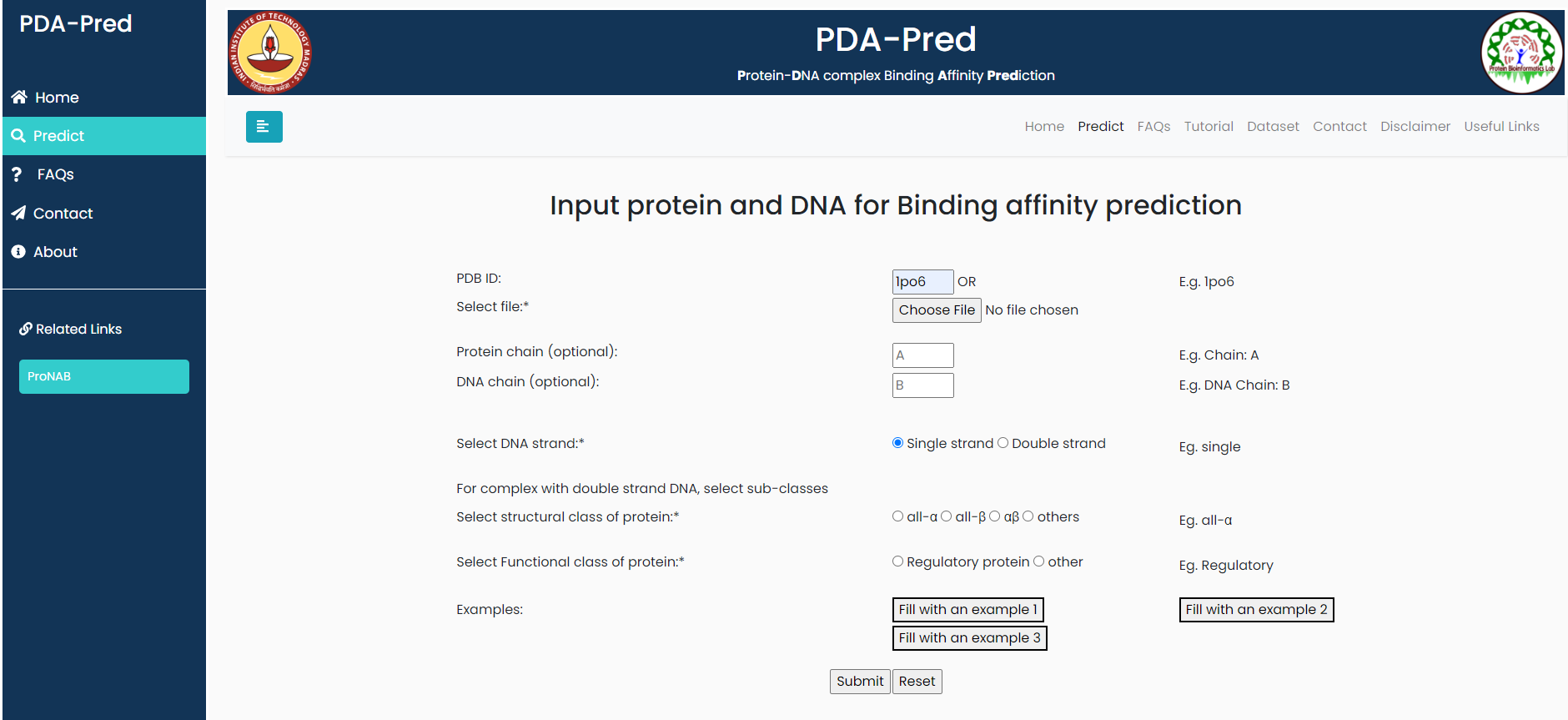
Crystal Structure of UP1 Complexed With DNA
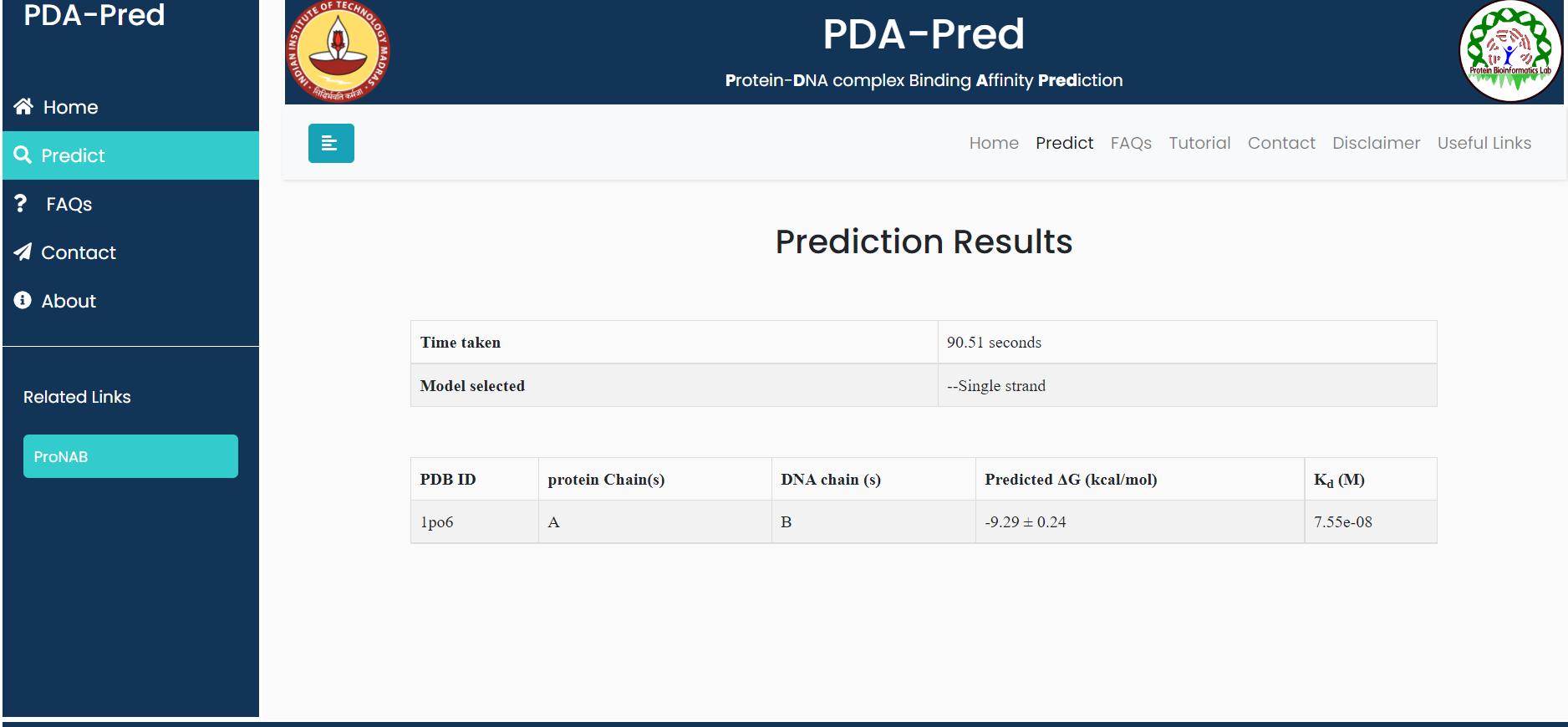
Input:PDB ID: 1po6, belongs to 'single strand DNA' classification.
Result for example 1:
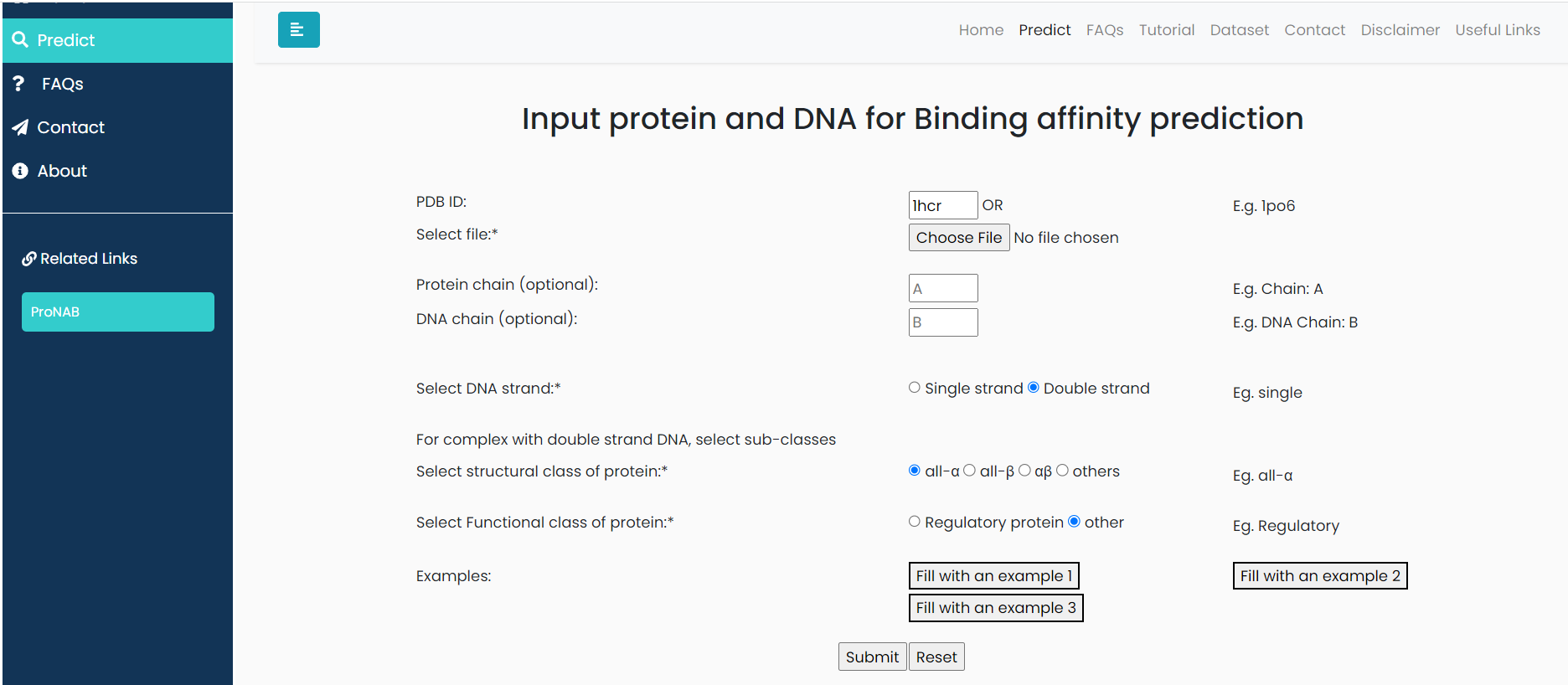
HIN RECOMBINASE BOUND TO DNA
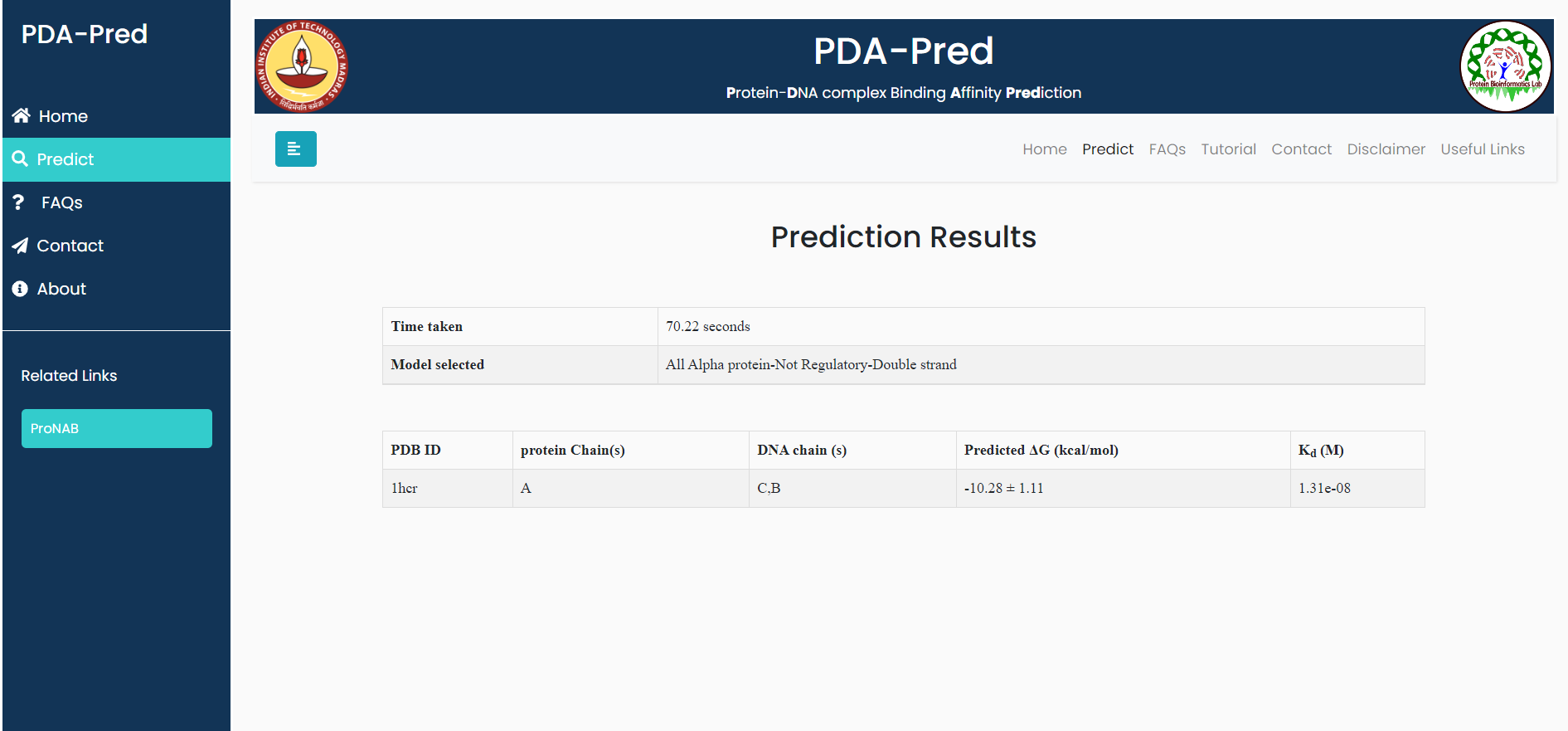
Input:PDB ID: 1hcr, Protein chain ID: A, DNA Chain ID: B,C, belongs to 'ds-All Alpha-non-regulatory' classification.
Result for example 2:
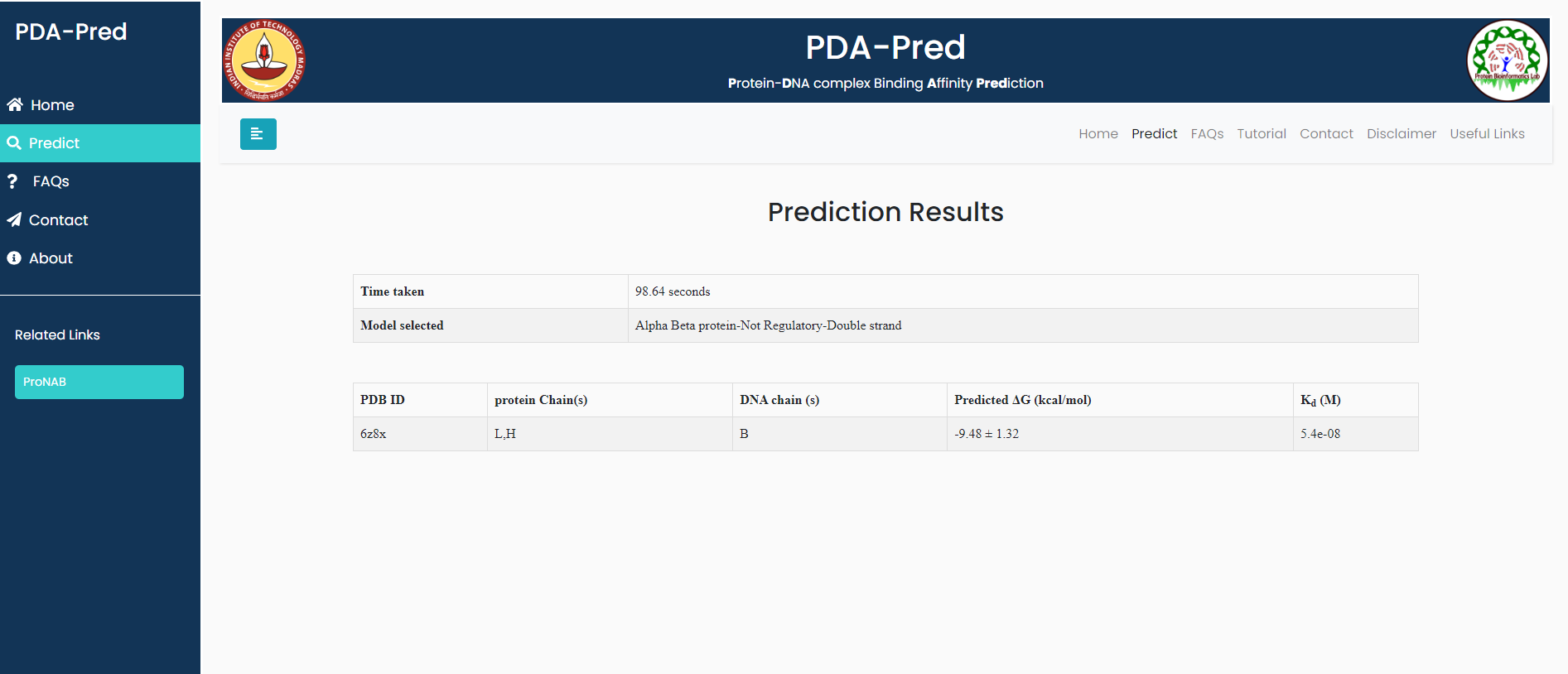
Complex of human alpha thrombin with DNA aptamer
Input: PDB: 6z8x,belongs to 'ds-Alpha-beta-non-regulatory'.
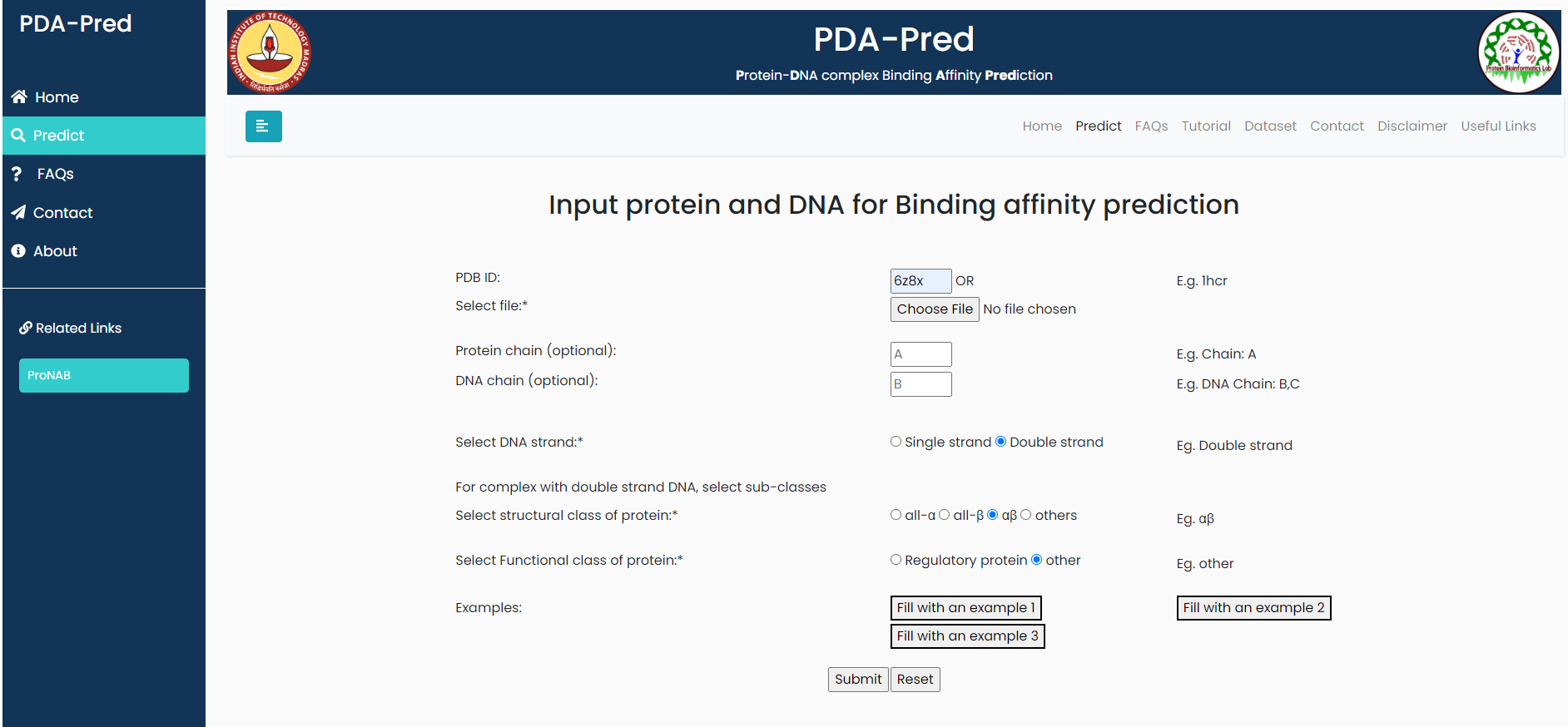
Result for example 3: