RNA SEQUENCE AS INPUT
The nucleotide sequence of the RNA target of interest must be provided as input in the FASTA format to the prediction form. The FASTA file format begins with a header, containing a ">" symbol followed by a name for the sequence. This header line is followed by the nucleotide sequence of the RNA.
Examples:
a) >HIV-1 TAR RNA
GGCAGAUCUGAGCCUGGGAGCUCUCUGCC
b) >Yeast phenylalanyl t-RNA
GCGGAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCA
SMALL MOLECULE INPUT FORMATS
The small molecule can be provided as input to the prediction form in both 1D and 3D formats. Currently the form accepts Simplified Molecular Input Line Entry System (SMILES) strings for the 1D format, and the Structural Data File (SDF) for the 3D format.
It is notable that the 1D SMILES strings for RNA-targeting small molecules is already available for each R-SIM database entry. To get the SDF file format for the corresponding small molecule, standard databases such as PubChem or ChemSpider can be used. Stand-alone cheminformatics toolkits such as OpenBabel or RDKit can also be used to interconvert between 1D and 3D file formats.
Examples:
| Molecule | SMILES format (1D) | SDF format (3D) |
|---|---|---|
| Benzene | c1ccccc1 | |
| Quercetin | C1=CC(=C(C=C1C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O)O)O | |
| Malachite green | CN(C)C1=CC=C(C=C1)C(=C2C=CC(=[N+](C)C)C=C2)C3=CC=CC=C3.[Cl-] |
RNA CATEGORIES IN RSAPred
RSAPred includes binding affinity prediction models for six RNA categories namely: Aptamers, miRNAs, Repeats, Ribosomal RNAs, Riboswitches, and Viral RNAs. Specifically, the viral RNA category includes a generic model trained on all viral RNA targets, and a HIV trans-activation response (TAR) element-specific model. This tutorial aims to enable users to decide the RNA category which might be suitable for their RNA target of interest. By selecting the corresponding model in the prediction form, users can get the predicted binding affinity values upon submission. The following table summarizes the RNA targets considered under each RNA category for model development.
| Aptamers | miRNAs | Repeats | Ribosomal RNAs | Riboswitches | Viral RNAs |
|---|---|---|---|---|---|
Cofactor-binding aptamers (Riboflavin, Biotin, NMN, Cyanocobalamin, FMN, Lysine, TPP etc.) and De novo designed aptamers with specific secondary structures |
Pre-, Pri-, and mature-miRNAs (includes both wild-type and mutant sequences) |
Disease-associated repeat regions (FXTAS, ALS, Huntington's, Fragile-X E-syndrome, Alzheimer's etc.) and G-quadruplex sequences |
Different regions of the bacterial ribosomal RNA (Peptidyl Transferase Center (PTC), 50S rRNA, 23S rRNA, decoding region (A-site) and Helix 22) |
All riboswitch sequences available in R-SIM database |
Viral IRES sequences, HIV RNA elements (TAR, FSS, RRE and PAS), Influenza virus promoter region, SARS-CoV-2 -1RF pseudoknot, EV71 SL-II RNA, Coxsackie virus B3 RNA and Polio virus loop B construct |
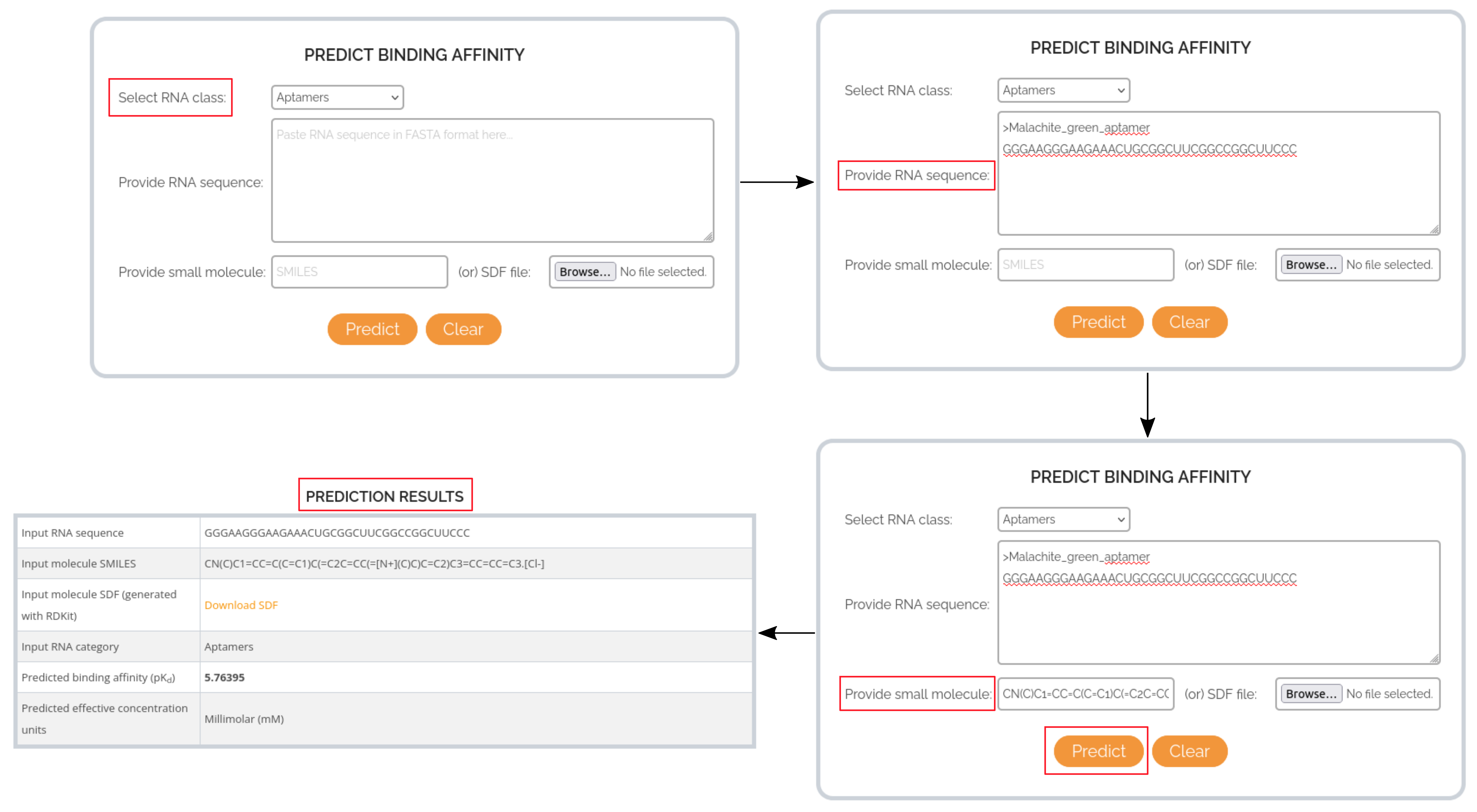
CASE STUDY 1: PREDICTING THE BINDING AFFINITY OF MALACHITE GREEN TO MALACHITE GREEN-BINDING RNA APTAMER
In this case, the RNA target of interest is the Malachite green-binding aptamer and the small molecule is Malachite green. From the R-SIM entry 385 the experimental binding affinity value for this RNA-small molecule interaction was found to be 5E-8 M (Kd). The corresponding log-scale binding affinity value (pKd) is 7.30. The following images describe how to get the predicted binding affinity for this interaction from RSAPred.
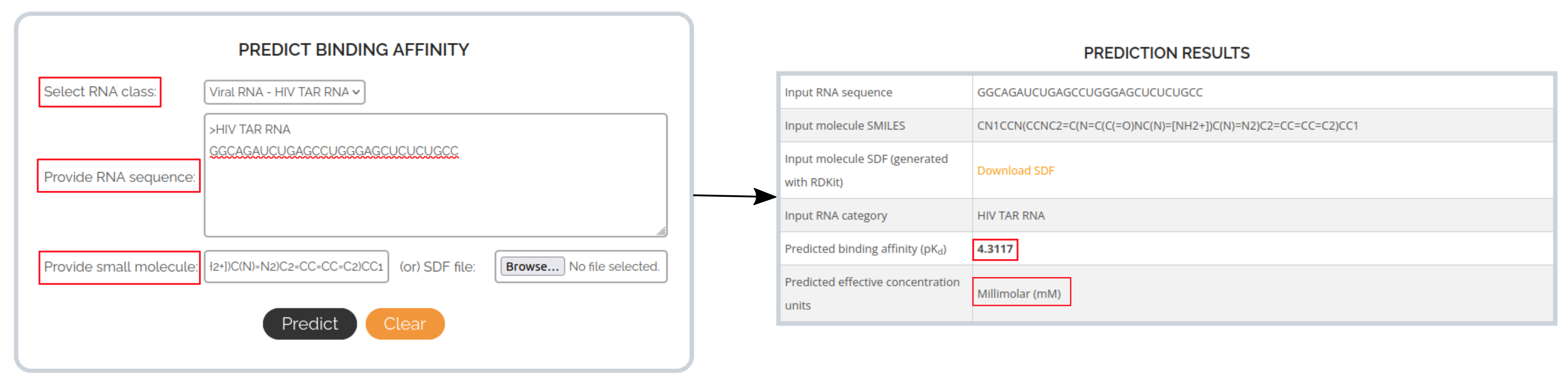
CASE STUDY 2: PREDICTING THE BINDING AFFINITY OF DMA-187 TO HIV-1 TAR RNA
In this case, the RNA target of interest is the HIV-1 transactivation response (TAR) element and the small molecule is a dimethyl amiloride derivative (DMA-187). From a recent QSAR study on HIV-1 TAR RNA, the experimental log-scale binding affinity value (pKd) for this RNA-small molecule interaction was found to be 4.634. The following images describe how to get the predicted binding affinity for this interaction from RSAPred.
INTERPRETING PREDICTION RESULTS
For the sample prediction results shown in case study 1, the interpretation can be done as follows:
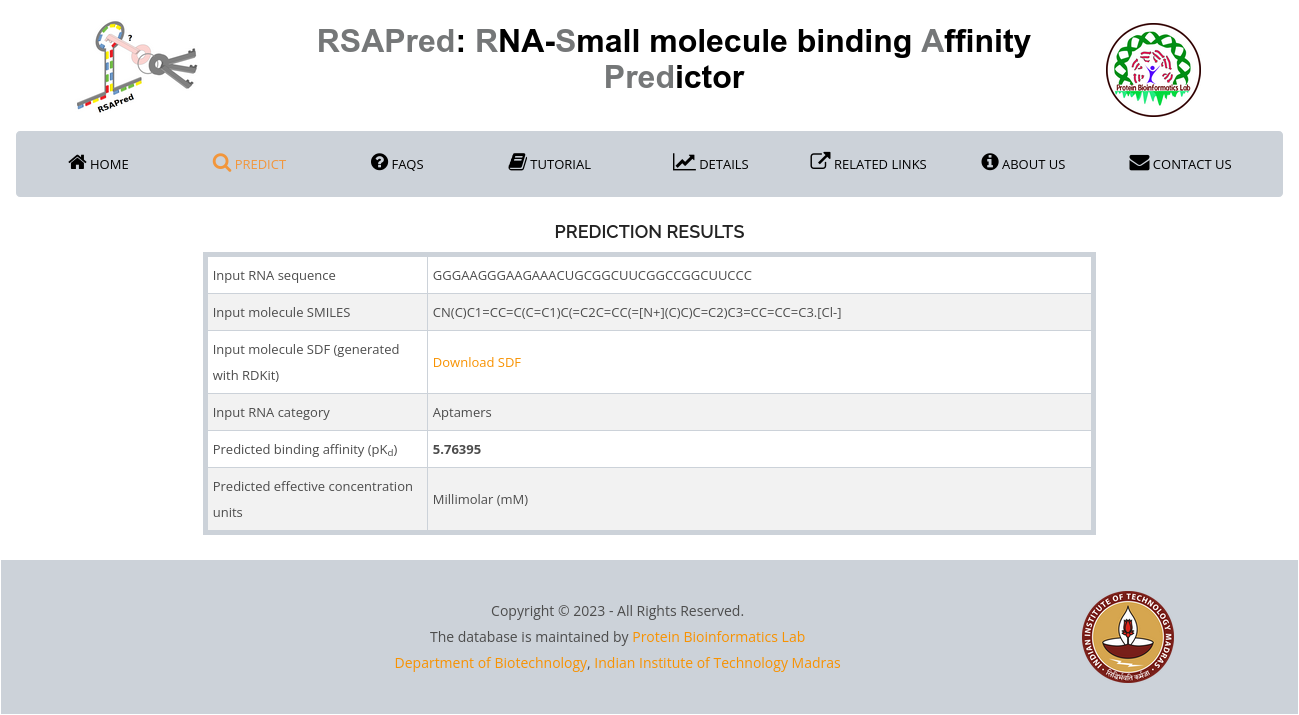
- The first four rows of the results table are used to recapitulate the user inputs received from the prediction form. They include the input RNA sequence, small molecule SMILES and SDF, and the RNA category chosen by the user.
- Predicted binding affinity (pKd): The log-scale dissociation constant (pKd) value predicted by the Aptamer-specific model in RSAPred.
- Predicted effective concentration units: The concentration unit corresponding to the pKd value predicted by the model. This is the predicted minimal concentration from which the small molecule is expected to be active against the RNA target of interest. The following table can be used as a reference to assign the predicted pKd values from RSAPred to their corresponding concentration units.
| pKd lower range (Inclusive) | pKd upper range (Exclusive) | Effective concentration units |
|---|---|---|
- |
3 |
Above millimolar (Likely inactive) |
3 |
6 |
Millimolar (mM) |
6 |
9 |
Micromolar (µM) |
9 |
12 |
Nanomolar (nM) |
12 |
- |
Sub-nanomolar (Highly active) |
|
© 2023, Protein Bioinformatics Lab, Indian Institute of Technology Madras All Rights Reserved. This server is maintained by Department of Biotechnology (IIT-M) |
 |