|
PCA-PredProtein-Carbohydrate complex binding Affinity Prediction |
 |
PCA-Pred
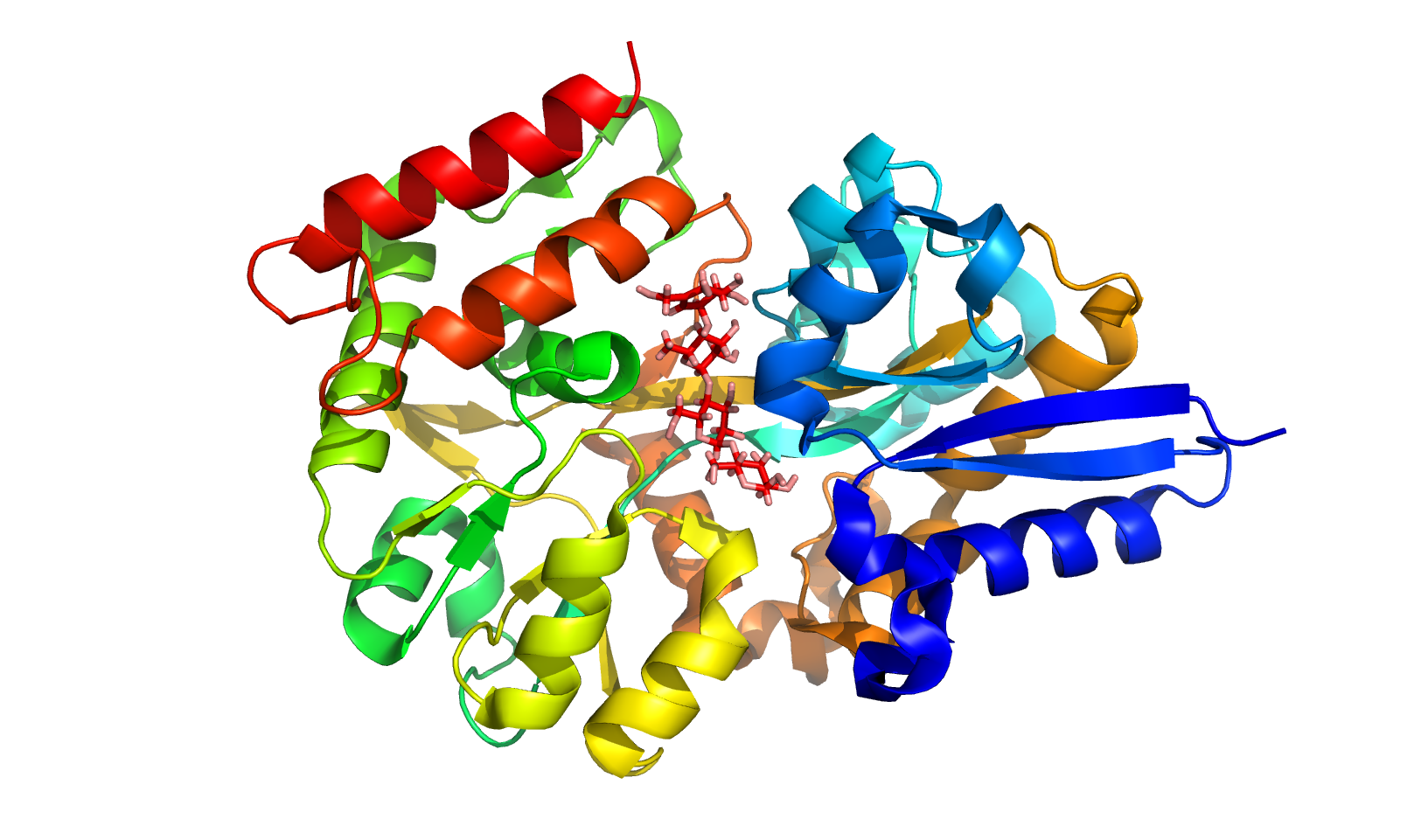
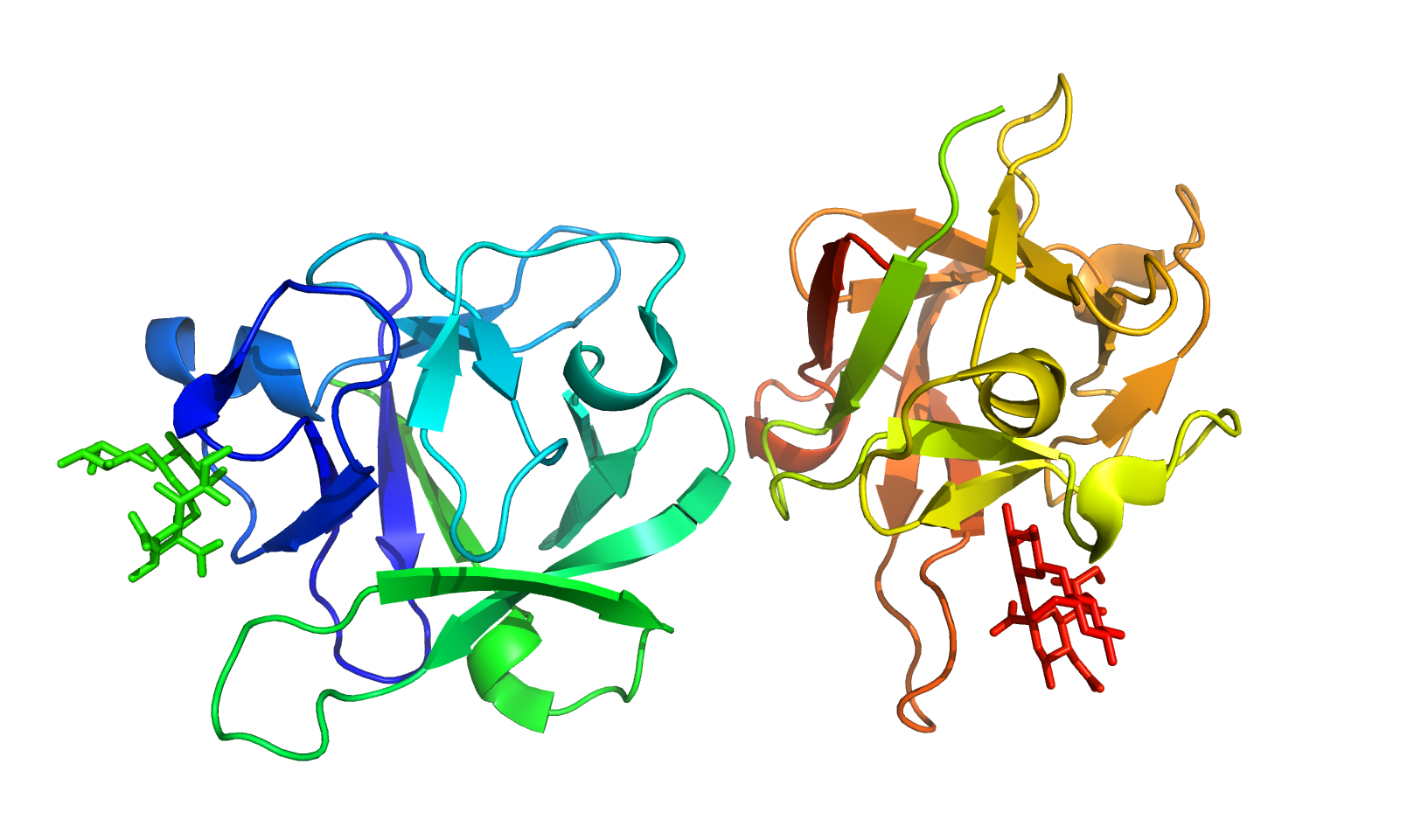
This server predicts protein-carbohydrate binding affinity using structure-based features and classes based on protein chains and saccharide count. Generally, binding site residues, accessible surface area, interactions between various atoms and their energy contributions are important to understand the binding affinity. PCA-Pred shows a correlation of 0.731 and a mean absolute error (MAE) of 1.149 kcal/mol in jack-knife test. In the test dataset, the performance remains consistent with a correlation of 0.723 with MAE of 1.158 kcal/mol.
Please click here to start prediction.