|
PCA-PredProtein-Carbohydrate complex binding Affinity Prediction |
 |
Tutorial
The predict page can be accessed from here.
This page takes input of protein-carbohydrate complex and predicts the binding affinity.
Click here to watch the video demonstration of the server
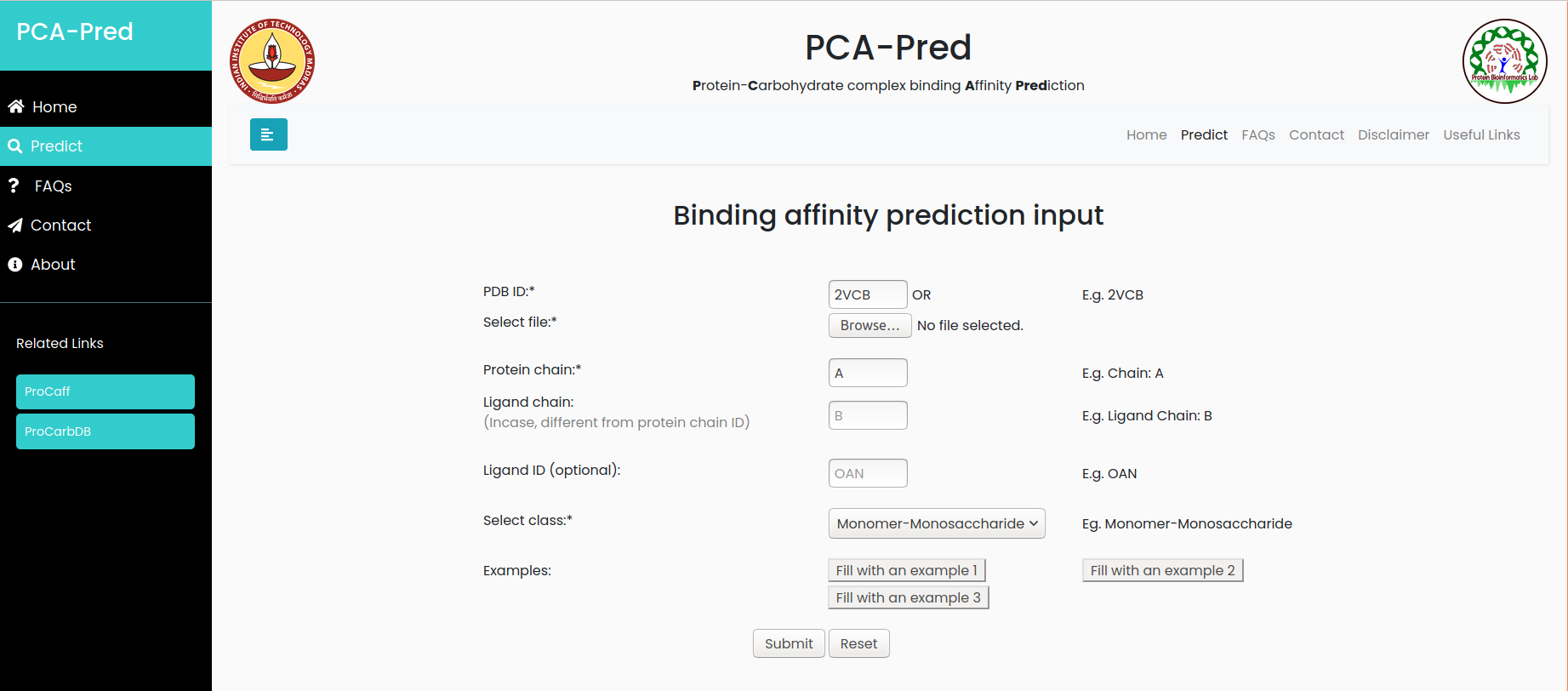
Input options:
- The page has option to enter PDB ID of protein-carbohydrate complex or user can upload their file in PDB format. *
- User has to enter the chain ID of protein. Multiple chain Ids can be entered without space or comma. (Eg. Chain ID: AB) *
- User has to enter ligand chain ID if carbohydrate chain ID is differenet from protein chain ID. (Optional)
- User can also enter specific ligand ID as mentioned in the PDB file.
User can also enter the multiple ligand ids separated by comma, without space inbetween and HETATM position is not required (Optional) - User should select one of the six classes for the prediction. *
- Monomer-Monosaccharide
- Dimer-Monosaccharide
- Oligomer-Monosaccharide
- Disaccharide
- Trisaccharide
- Oligosaccharide
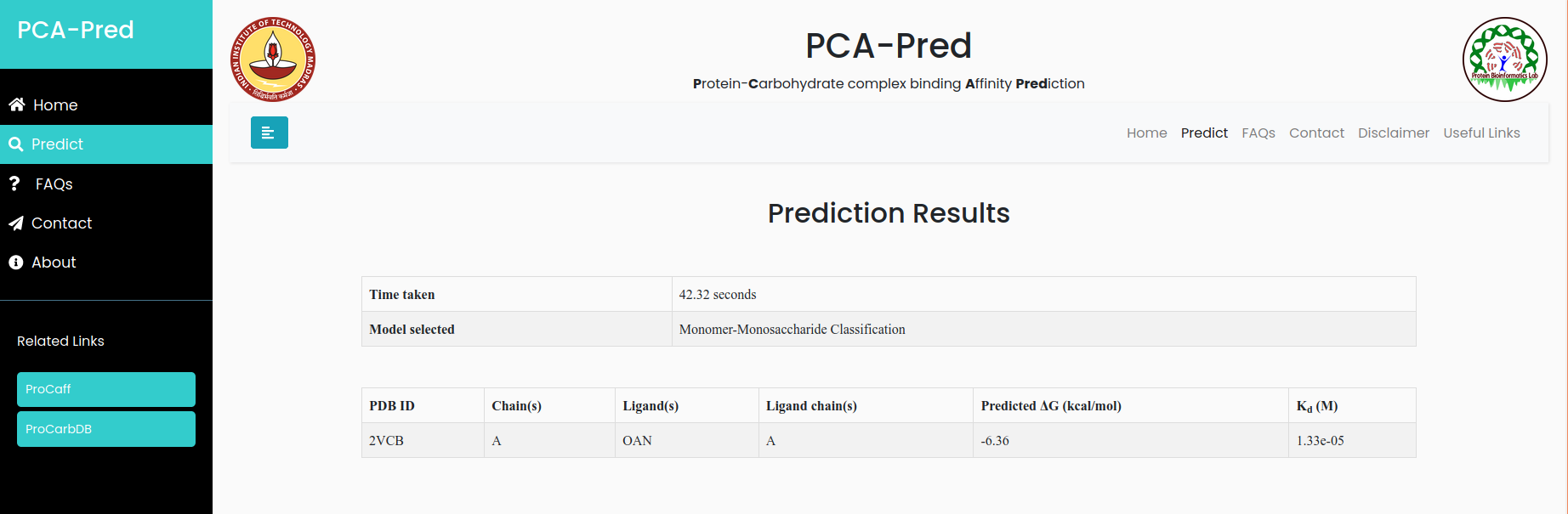
Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc
Input:PDB ID: 2VCB, Chain ID: A, belongs to 'Monomer-Monosaccharide' classification.
Result for example 1:
Human Galectin-3 CRD in complex with Lactose
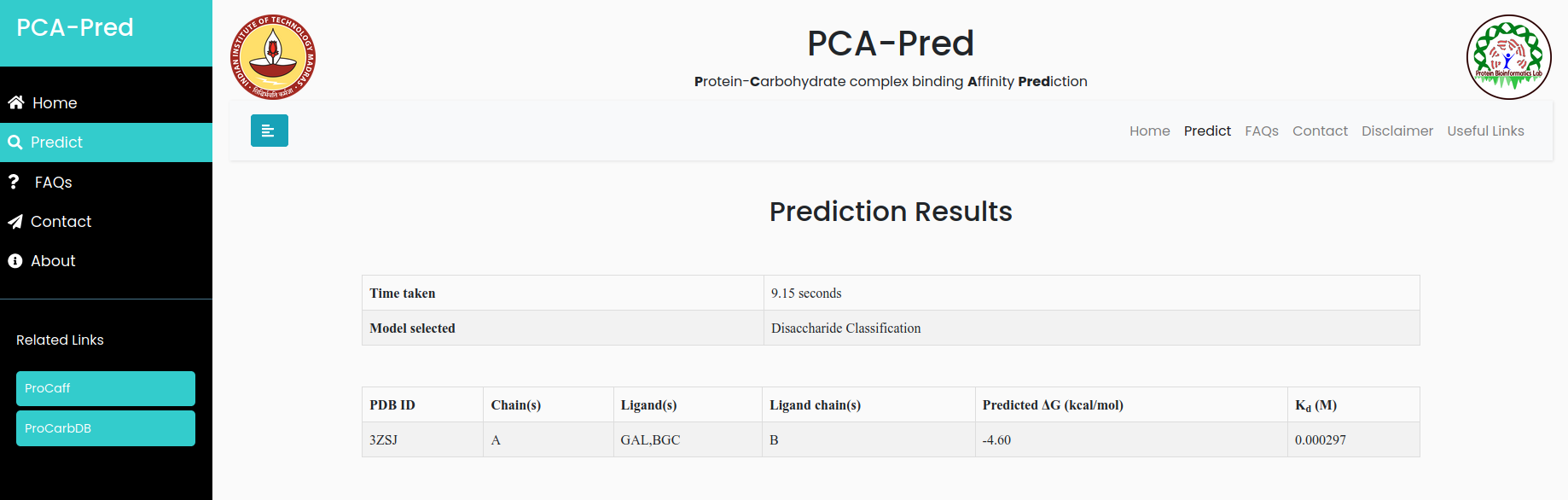
Input:PDB ID: 3ZSJ, Protein chain ID: A, Carbohydrate Chain ID: B, belongs to 'Disaccharide' classification.
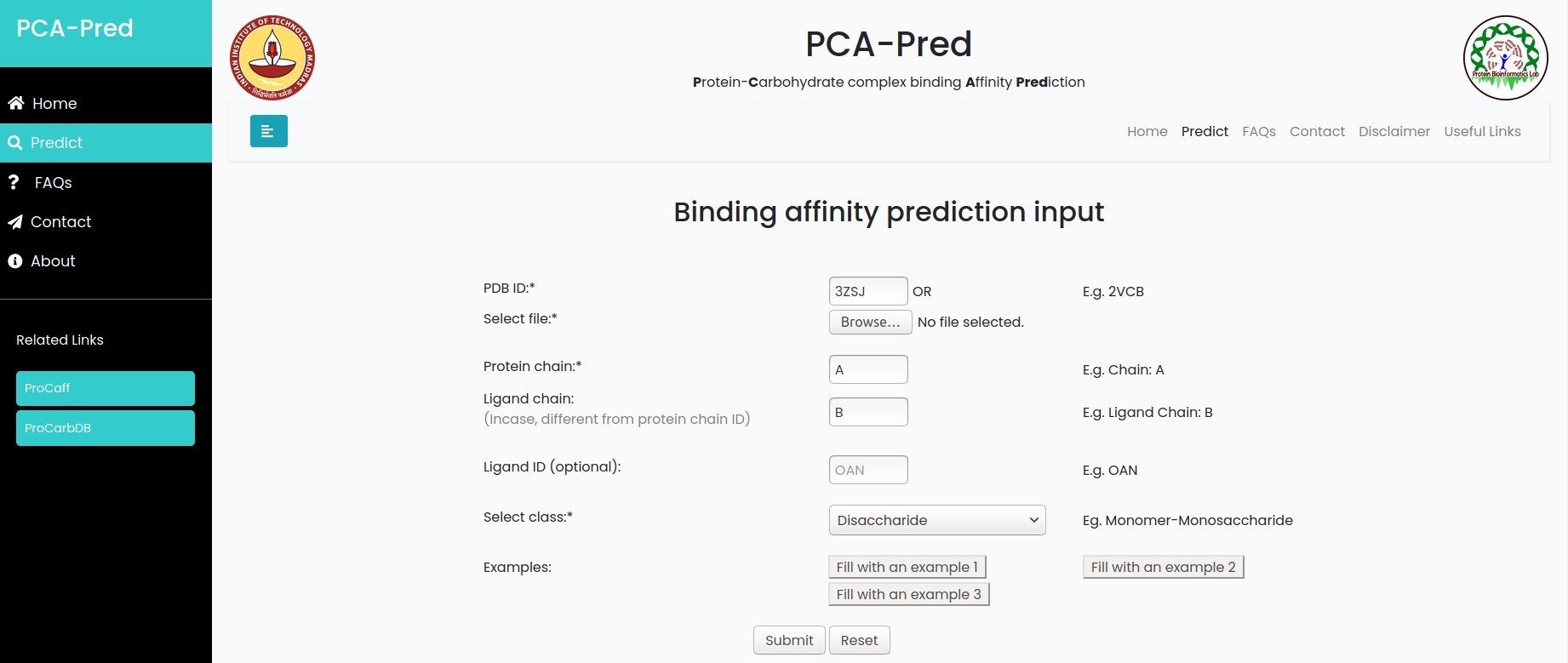
Result for example 2:
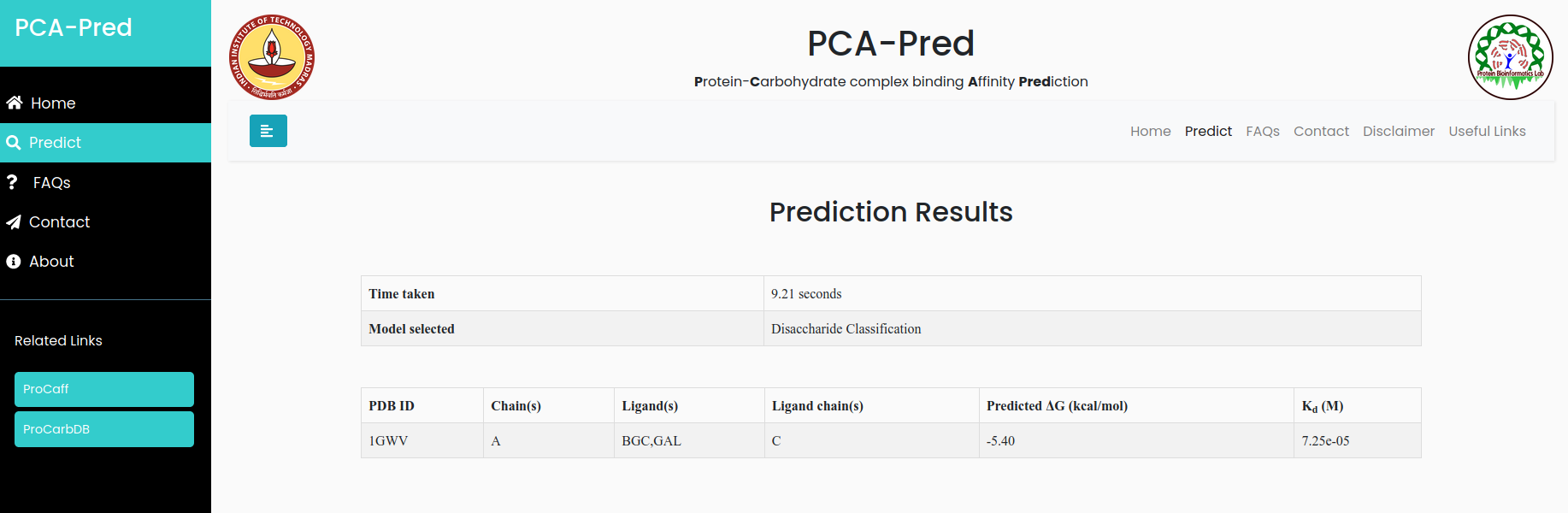
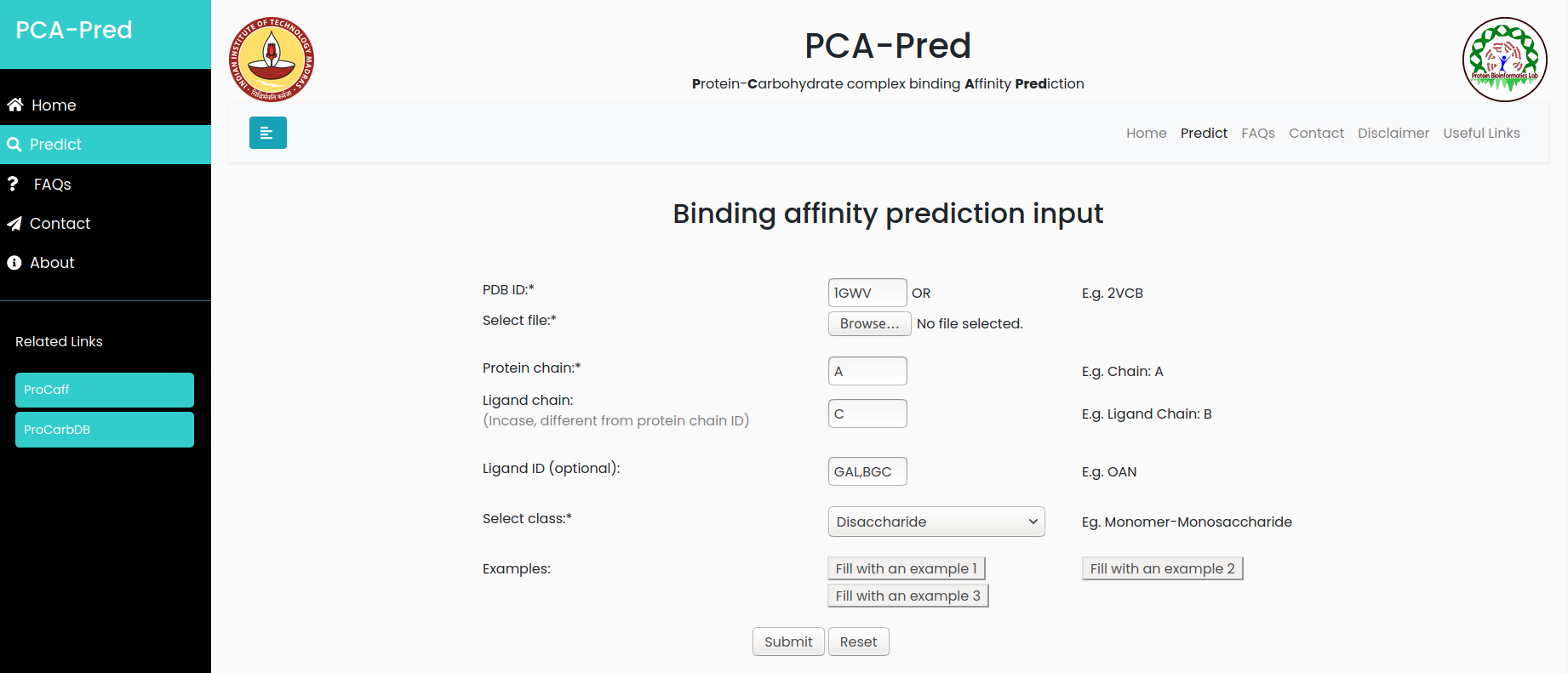
Alpha-1,3 Galactosyltransferase-Lactose Complex of Bos taurus
Input:PDB ID: 1GWV, Protein chain ID: A, Carbohydrate Chain ID: C, Ligand ID:GAL, BGC and it belongs to 'Disaccharide' classification.
Result for example 3: