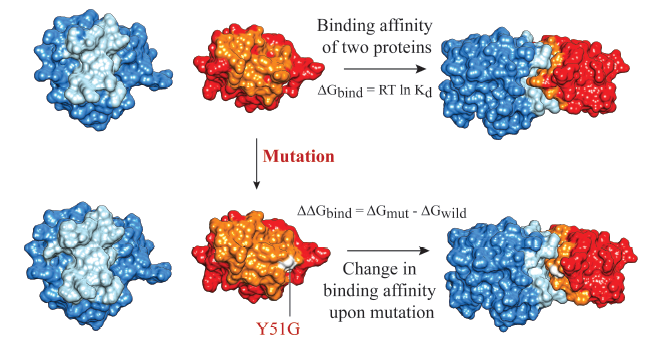
Binding Affinity of Biomolecular Complexes
Our research focuses on studying the binding affinity of various protein-based complexes, including interactions with other proteins, nucleic acids, and carbohydrates. We aim to understand the molecular and structural factors that influence binding strength and specificity. We curate and develop specialized databases of experimentally characterized biomolecular interactions. Using these datasets, we build computational models to predict binding affinity for both wild-type complexes and mutation-induced changes in affinity. We aim to advance mechanistic understanding of molecular recognition and provide user-friendly resources that support the broader research community.
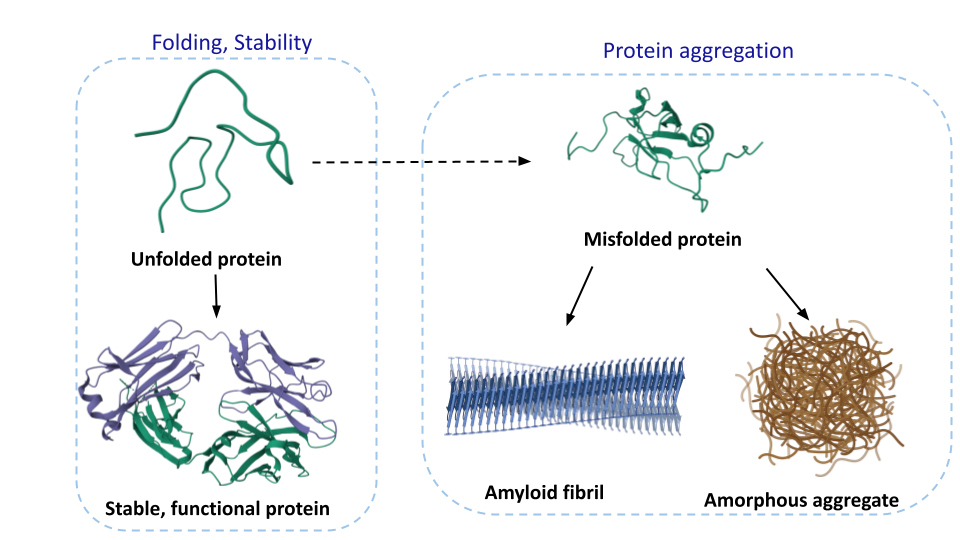
Protein folding, Stability, and Aggregation
Protein folding is a fundamental biophysical process that governs how polypeptide chains acquire their native three-dimensional structures essential for biological function. Computational algorithms leveraging high-resolution structural data from cryo-EM, and X-ray crystallography provide unprecedented insights into folding pathways, energy landscapes, and stability determinants. Our curated databases compile experimental data on protein thermodynamics from diverse biophysical techniques, as well as the mechanistic and kinetic aspects of protein aggregation. These resources enable systematic analysis of folding-stability relationships and their implications for protein function and disease.
- Creating and maintaining curated databases of thermodynamic data of proteins, mechanistic and kinetic data on protein aggregation
- Developing tools for the prediction of stability changes upon mutations
- Engineering tools for prediction of aggregation prone regions, and aggregation kinetics in wild type and mutant proteins

Structure-based Drug Design
Structure-Based Drug Design (SBDD) is a rational, computation-driven approach that uses three-dimensional structures of biological targets to guide drug discovery. By analyzing atomic-level features of proteins and their complexes, we design small molecules with improved specificity and affinity. Our work combines experimental structural data with molecular docking, molecular dynamics simulations, and free-energy calculations. We focus on how conformational changes and mutations influence ligand binding. This framework supports informed design choices based on molecular-level insights. Overall, our aim is to translate structural understanding into practical tools that accelerate drug discovery.
- Combining experimental structures with computational methods to model ligand–target interactions
- Understanding the impact of protein flexibility and mutations on binding behavior
- Developing predictive models and streamlined pipelines for efficient lead identification

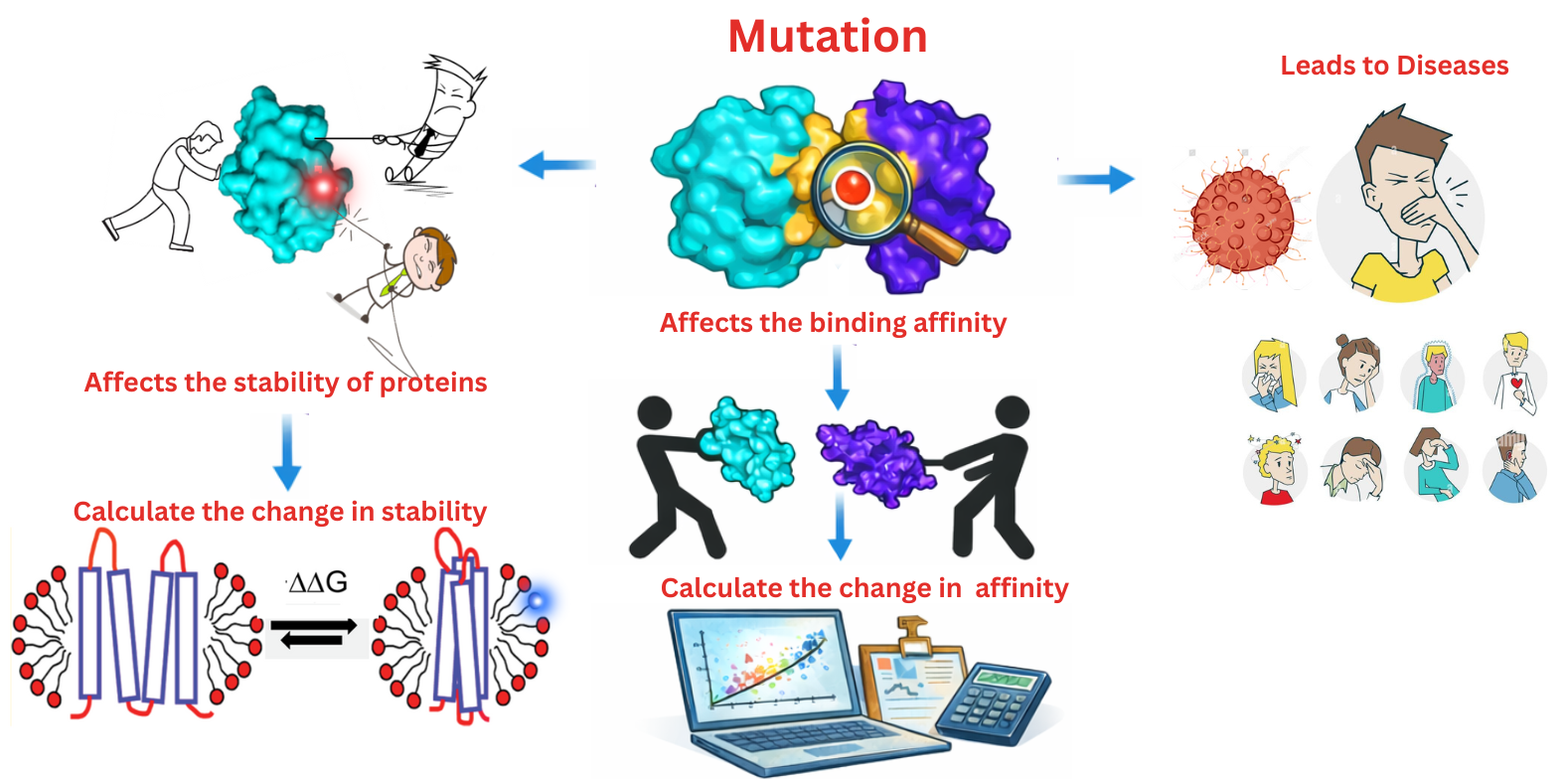
Disease Causing Mutations
A disease-causing mutation is a change in the DNA of a gene that increases a person's risk for a genetic disorder or disease, such as cancer. These mutations can be inherited or acquired during a person's life. Identifying a disease-causing mutation may help with the prevention, diagnosis, and treatment of diseases, but it's important to remember that not everyone with such a mutation will actually develop the disease. This is also known as a pathogenic variant, predisposing mutation, or susceptibility gene mutation. Our lab is dedicated to identifying and understanding disease-causing mutations, with a special focus on missense mutations in proteins. We investigate how these mutations alter protein structure, which can lead to the development of diseases.
- Understanding Mutation Impact: We study how mutations affect protein structure and function.
- Identifying Key Features: We aim to pinpoint the specific protein features that are disrupted by these mutations
- Developing Predictive Tools: We build databases and leverage machine learning and deep learning to create computational models for predicting and identifying disease-causing mutations.
Membrane Proteins
Transmembrane proteins (TMPs) interact with lipid bilayer of cell membranes and perform several salient biological functions. These cellular functions are mainly dictated by their structures. The folding, stability and functions of TMPs are governed by the insertion of secondary structural elements into the cell membrane followed by interactions with other proteins, lipid bilayer and aqueous environment. Our research focuses on understanding the stability, binding affinity, and disease state implications of mutations in membrane proteins. Our primary research objectives are:
- Identifying the important features governing the membrane protein stability as well as developing models for predicting the stability of membrane proteins
- Predicting the binding affinities of the membrane protein–protein complexes using multiple regression-based technique
- Large-scale analysis of disease-causing and neutral mutations in human membrane proteins using computational approaches

Multiomics Analysis
Next-generation sequencing is a rapidly advancing high-throughput technology that determines the sequences of an organism's genetic material. This helps us to study the genetic changes associated with disease conditions. Multi-omic analysis is an integrative approach that combines data from multiple layers of biological regulation, including genomics, transcriptomics, epigenetics, proteomics, and metabolomics. This systems-level approach helps in understanding complex biological interactions, providing deeper insights into disease mechanisms, their progression, and the identification of potential therapeutic targets.
- Developing personalized treatment strategies for diseases like cancer, neurodegenerative diseases, etc., using the next-generation sequencing data.
- Analysing the genomics data to understand the effect of mutations in cancer and to identify population-specific biomarkers
- Integrating the bulk and single-cell RNA sequencing to understand the tumour heterogeneity in cancer and for neoantigen prediction.
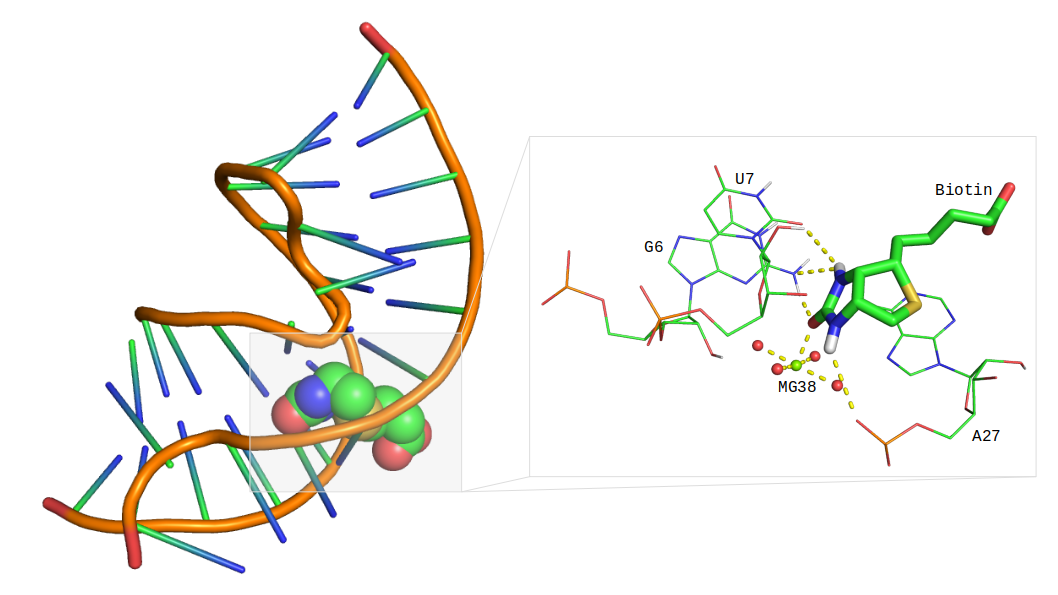
RNA-targeted Drug Discovery
Ribonucleic Acids (RNAs) are structured and functional molecules with versatile roles in cellular regulation, making them potential targets for drug development. Currently, a large fraction of proteins remain undruggable due to their essential nature. To address this growing need for identification of novel targets in drug discovery, we have closely studied the potential of RNAs to act as druggable targets. Our primary research objectives are:
- To develop and maintain a comprehensive database of RNA-small molecule interactions with binding affinity and experimental information.
- Accurate prediction of RNA-small molecule binding affinity using machine learning
- Development of methods for RNA target identification and binding site prediction